Files and File Types
The primary file types you’ll see related to DNA sequence analysis are:
- fasta
- fastq
- gtf/gff
- sam/bam/cram
- vcf
Sequence based file types
Sequence based files first started out as fasta with paired qual files (Sanger and 454), with Illumina and quality scores being used more, the fastq file became the default output from DNA sequencers. These days additional file types are being used, including fast5 by Oxford Nanopore and ‘unmapped’ bam files by Pacific Biosciences.

fasta
The fasta format uses the ‘>’ to indicate a new sequence followed by the name of the sequence on the same line. The following line(s) are the DNA sequence and may be split on multiple lines (wrapped), until the next ‘>’ is reached. Genome and transcriptome files are most often in fasta format.

Qual files are so rarely used these days and so are not discussed.
fastq
fastq files combine the sequence and quality scores into 1 file. Each sequence here has 4 lines (should be enforced strictly), header, sequence, historical ‘+’, and quality.

CASAVA 1.8 Read IDs
@EAS139:136:FC706VJ:2:2104:15343:197393 1:Y:18:ATCACG
- EAS139 the unique instrument name
- 136 the run id
- FC706VJ the flowcell id
- 2 flowcell lane
- 2104 tile number within the flowcell lane
- 15343 ’x’-coordinate of the cluster within the tile
- 197393 ’y’-coordinate of the cluster within the tile
- 1 the member of a pair, 1 or 2 (paired-end or mate-pair reads only)
- Y Y if the read fails filter (read is bad), N otherwise
- 18 0 when none of the control bits are on, otherwise it is an even number
- ATCACG index sequence
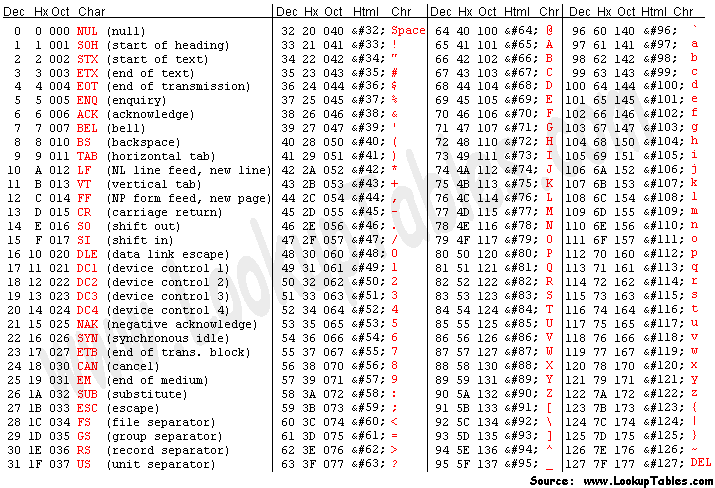
Quality scores
Quality scores are paired 1 to 1 with sequence characters.
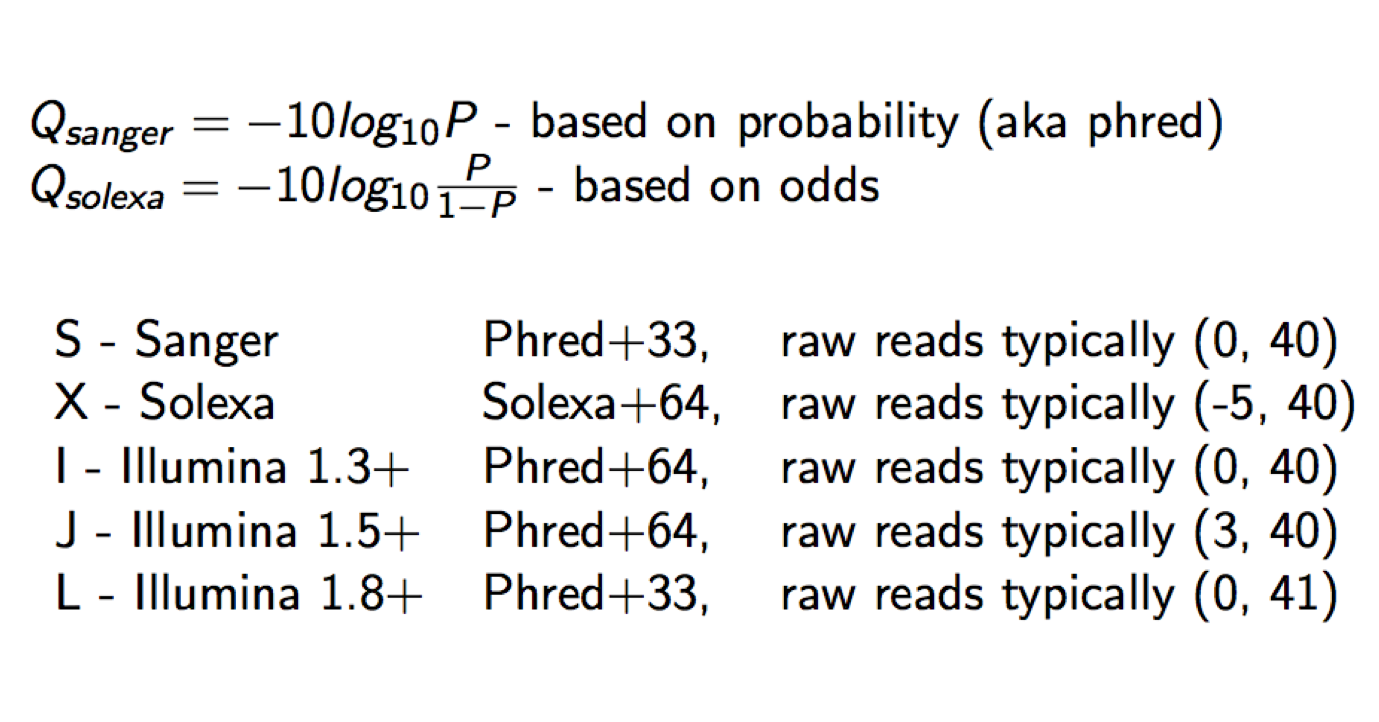
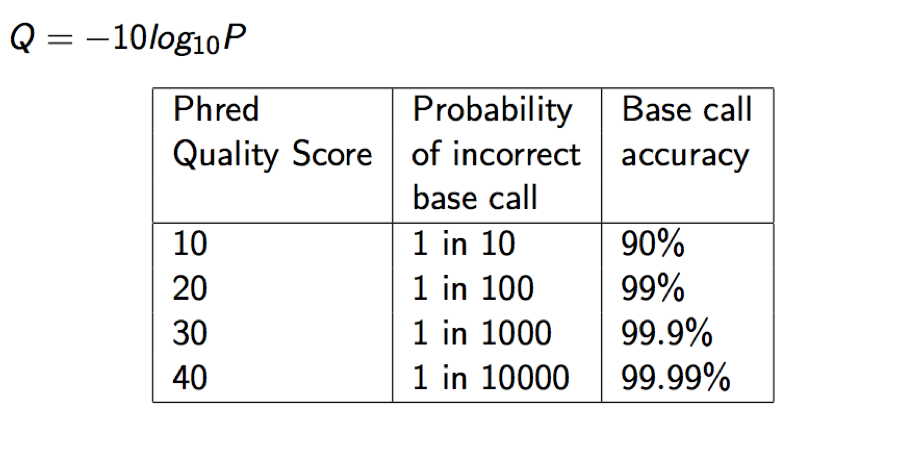
Each quality character has a numerical value associated with it (ASCII value). In Illumina 1.8+ you subtract 33 from the ascii value associated with the quality character to get the quality score.
Annotation based file Types
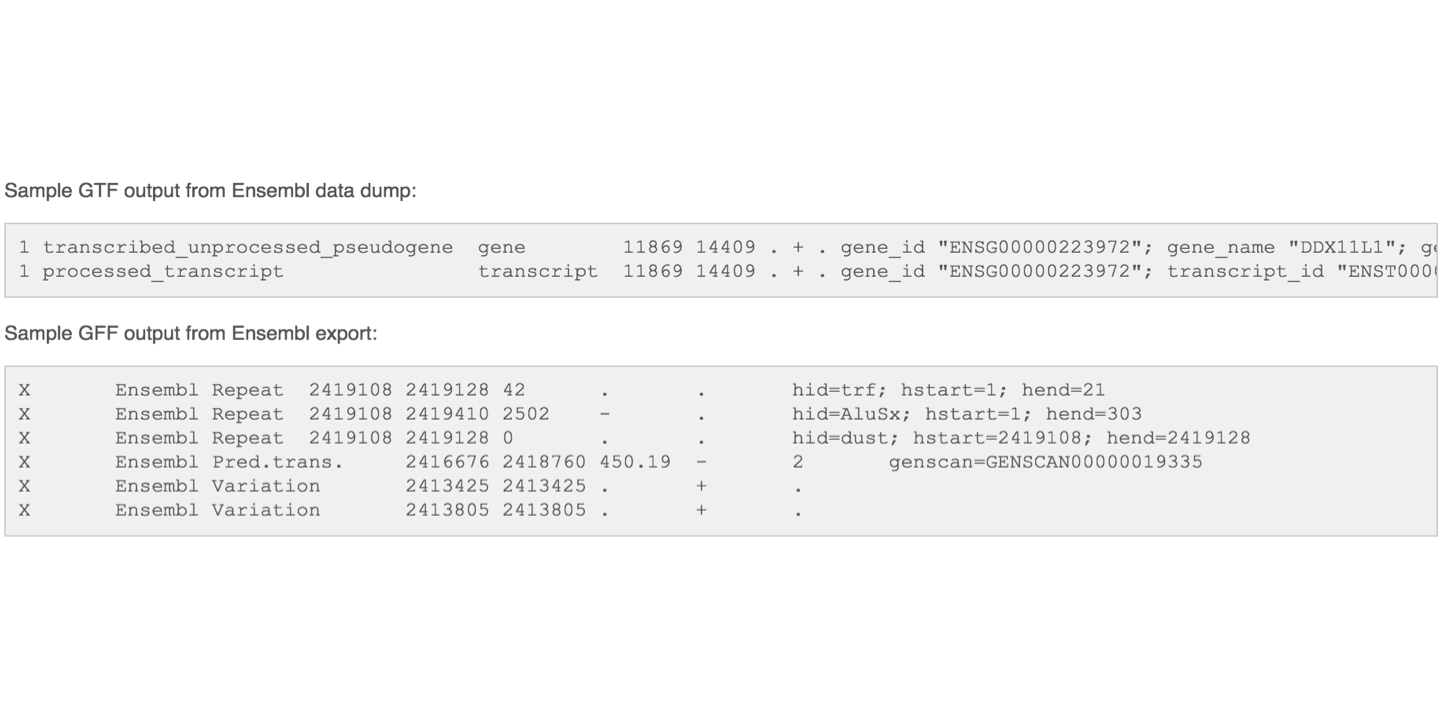
Gene Transfer Format (GTF) / Gene Feature Format (GFF)
Describes feature (ex. gene) locations within a sequence file (ex. genome).
The GFF (General Feature Format) format consists of one line per feature, each containing 9 columns of data (fields). The GTF (General Transfer Format) is identical to GFF version 2. The GTF file format tends to have stricter requirements and expectations on how feature descriptions are formated.
Fields must be tab-separated and all fields must contain a value; “empty” fields should be denoted with a ‘.’.
Columns: Seqname: Name of the sequence chromosome Source: the program, or database, that generated the feature
Feature: feature type name examples include gene, exon, cds, etc.
Start: start position of the feature sequences begin at 1
End: stop position of the featur Sequences begin at 1
Score: a floating point value (e.g. 0.01)
Strand: Defined as ‘+’ (forward),or ‘-’ (reverse)
Frame: One of ‘0’, ‘1’, ‘2’, ‘0’ Represents the first base of a codon.
Attribute: A semicolon-separated list of tag-value pairs Providing additional information about each feature.
Sources
- Ensembl genomes ( http://ensemblgenomes.org/ ) and annotation at Biomart ( http://www.ensembl.org/biomart/martview/ )
- Human/mouse: GENCODE (uses Ensembl IDs) ( https://www.gencodegenes.org/ ), but may need some manipulation to work with certain software.
- NCBI genomes ( http://www.ncbi.nlm.nih.gov/genome/ )
- Many specialized databases (Phytozome, Patric, VectorBase, FlyBase, WormBase)
Alignment based file types
SAM/BAM/CRAM
The project managing the specifications and common software is http://www.htslib.org/.
SAM (Sequence Alignment/Map) format is the unified format for storing read alignments to a reference sequence (consistent since Sept. 2011). http://samtools.github.io/hts-specs/SAMv1.pdf http://samtools.github.io/hts-specs/SAMtags.pdf
BAM are compressed SAMs (binary, not human-readable). They can be indexed to allow rapid extraction of information, so alignment viewers do not need to uncompress the whole BAM file in order to look at information for a particular coordinate range, somewhere in the file.
Indexing your BAM file, mybam.bam, will create an index file, mybam.bam.bai, which is needed (in addition to the BAM file) by viewers and some downstream tools.
CRAM is an even more compressed SAM/BAM file and has the following major objectives:
- Significantly better lossless compression than BAM
- Full compatibility with BAM
- Effortless transition to CRAM from using BAM files
- Support for controlled loss of BAM data
CRAM files are also indexed and produce .cram.crai index files.
The SAM format contain two regions:
The header section - Each header line begins with character ’@’ followed by a two-letter record type code.
The alignment section - Each alignment line has 11 mandatory fields. These fields always appear in the same order and must be present, but their values can be ’0’ or ’*’, if the corresponding information if unavailable, or not applicable.
Header section
Each header line begins with the character ‘@’ followed by one of the two-letter header record type codes. In the header, each line is TAB-delimited and, apart from @CO lines, each data field follows a format ‘TAG:VALUE’ where TAG is a two-character string that defines the format and content of VALUE.
The following tags are required:
- @HD The header line. The first line if present.
- VN* Format version. Accepted format: /^[0-9]+.[0-9]+$/.
- @SQ Reference sequence dictionary. The order of @SQ lines defines the alignment sorting order.
- SN* Reference sequence name. The SN tags and all individual AN names in all @SQ lines must be distinct. The value of this field is used in the alignment records in RNAME and RNEXT fields. Regular expression: [:rname:∧ =][:rname:]
- LN* Reference sequence length. Range: [1, 2 31 − 1]
- @RG Read group. Unordered multiple @RG lines are allowed.
- ID* Read group identifier. Each @RG line must have a unique ID. The value of ID is used in the RG tags of alignment records. Must be unique among all read groups in header section. Read group IDs may be modified when merging SAM files in order to handle collisions.
Many more tags are detailed in the specifications.
Alignment section
The alignment section consists of 11 tab delimited fields.
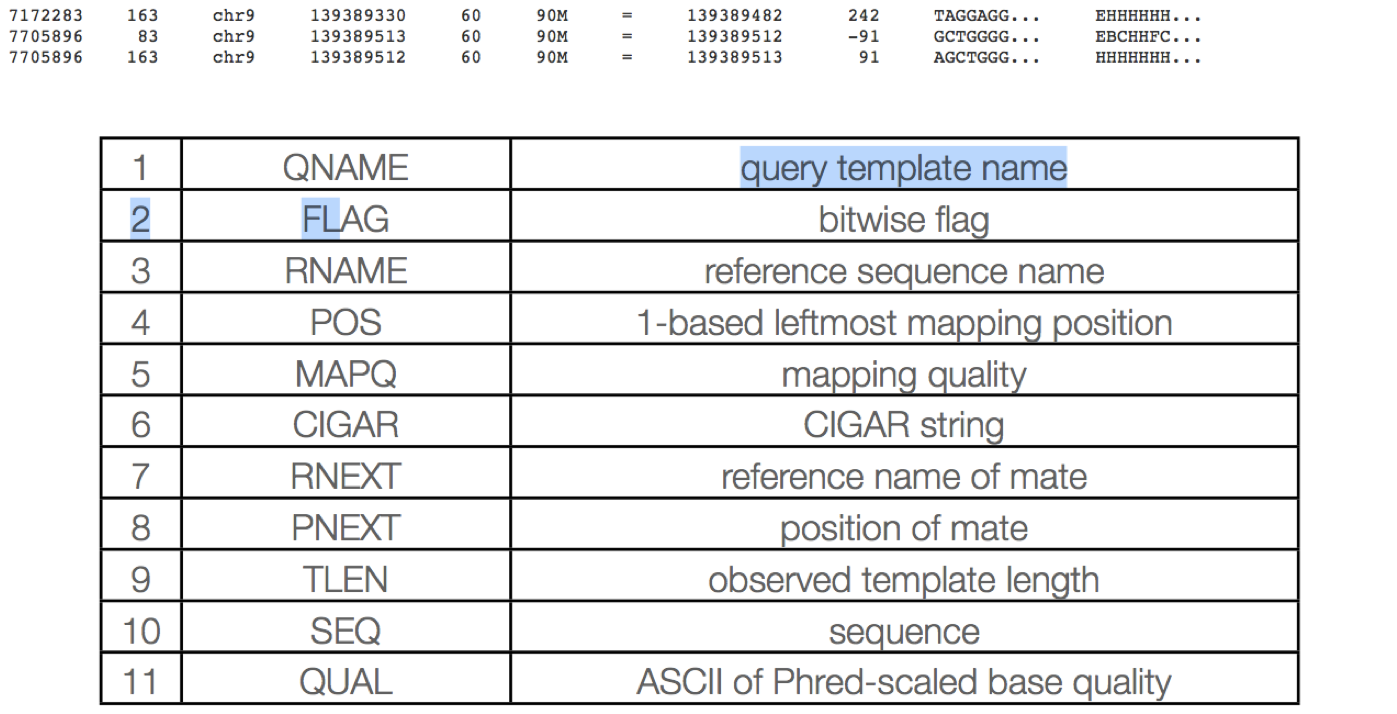
QNAME: Query name
Read IDs are truncated at first whitespace (spaces / tabs), which can make them non-unique. Illumina reads with older IDs have trailing “/1” and “/2” stripped (this information is recorded in the next field). Illumina reads with newer IDs have second block stripped (read number is recorded in the next field).
@FCC6889ACXX:5:1101:8446:45501#CGATGTATC/1 ⇒ @FCC6889ACXX:5:1101:8446:45501
@HISEQ:153:H8ED7ADXX:1:1101:1368:2069 1:N:0:ATCACG ⇒ @HISEQ:153:H8ED7ADXX:1:1101:1368:2069
FLAG: Alignment flags
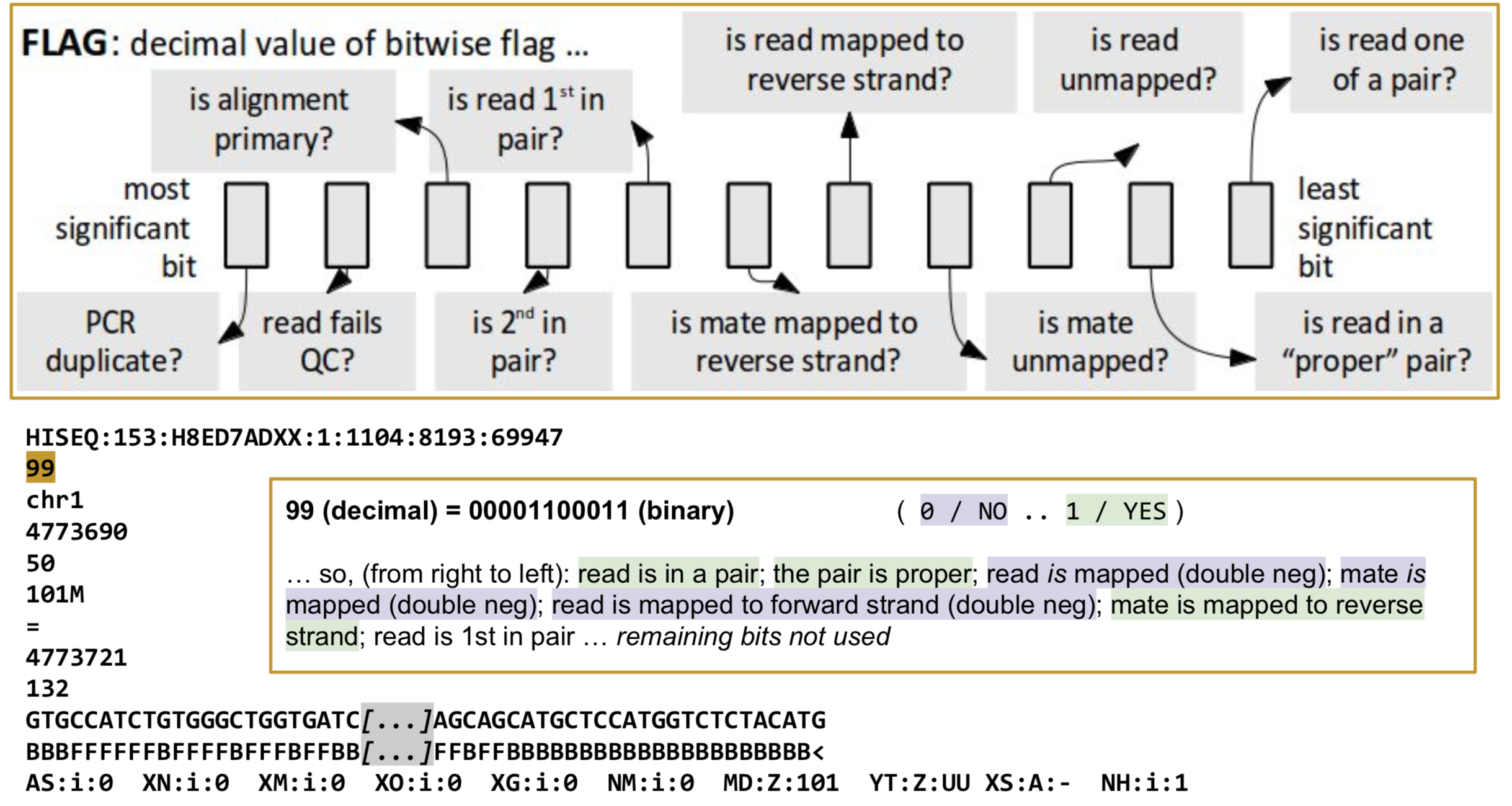
Tool to help explain flags
https://broadinstitute.github.io/picard/explain-flags.html
RNAME: reference sequence name
Reference sequence ID (from fasta header) matches a @SQ field in the header section.
POS: 1-based leftmost position of (post-clipping) aligned read
MAPQ: mapping quality (phred scaled)
MAPQ, contains the “phred-scaled posterior probability that the mapping position” is wrong.
In a probabilistic view, each read alignment is an estimate of the true alignment and is therefore also a random variable. It can be wrong. The error probability is scaled in the Phred. For example, given 1000 read alignments with mapping quality being 30, one of them will be incorrectly mapped to the wrong location on average.
A value 255 indicates that the mapping quality is not available.
MAPQ explained
The calculation of mapping qualities is simple, but this simple calculation considers many of the factors below:
- The repeat structure of the reference. Reads falling in repetitive regions usually get very low mapping quality.
- The base quality of the read. Low quality means the observed read sequence is possibly wrong, and wrong sequence may lead to a wrong alignment.
- The sensitivity of the alignment algorithm. The true hit is more likely to be missed by an algorithm with low sensitivity, which also causes mapping errors.
- Paired end or not. Reads mapped in proper pairs are more likely to be correct.
When you see a read alignment with a mapping quality of 30 or greater, it usually implies:
- The overall base quality of the read is good.
- The best alignment has few mismatches.
- The read has just one ‘good’ hit on the reference, which means the current alignment is still the best even if one or two bases are actually mutations, or sequencing errors.
In practice however, each mapping application seems to compute the MAPQ in their own way.
CIGAR: Compact Idiosyncratic Gapped Alignment Report (CIGAR)
The CIGAR string is used to calculate how a sequence aligns to the reference. Format: [0-9][MIDNSHP][0-9][MIDNSHP]… where M = match, I/D = insertion / deletion, N = skipped bases on reference (splicing), S/H = soft / hard clip (hard clipped bases no longer appear in the sequence field), P = padding.
e.g. “100M” means that all 100 bases in the read align to bases in the reference.

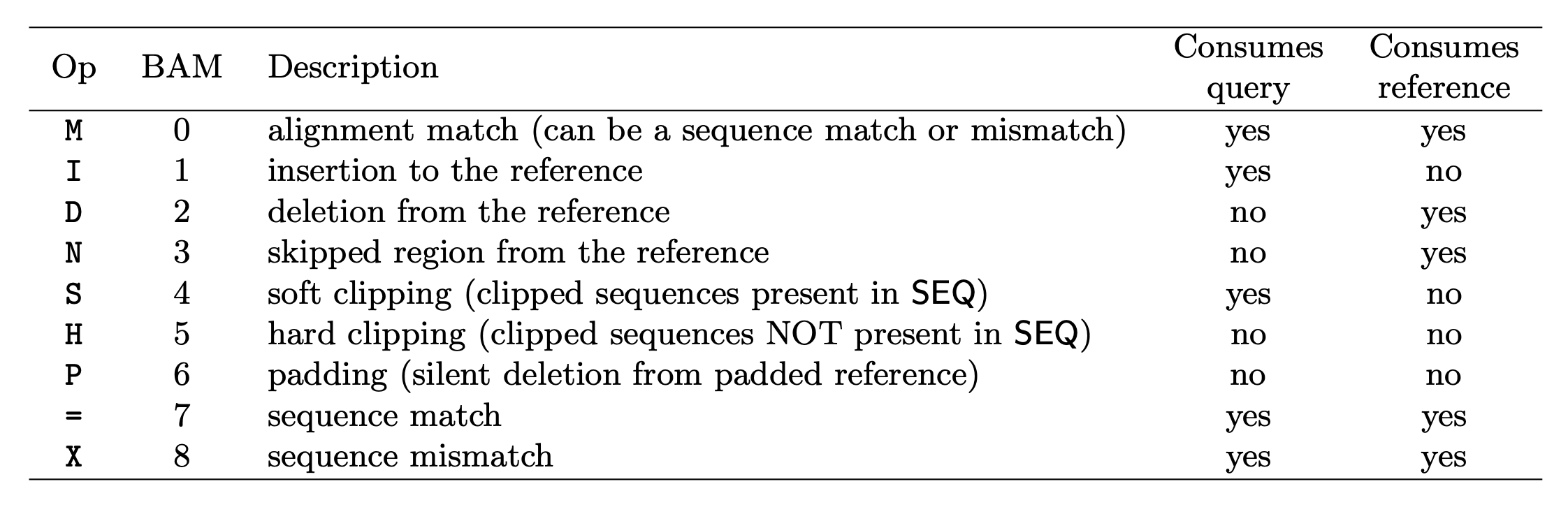
- “Consumes query” and “consumes reference” indicate whether the CIGAR operation causes the alignment to step along the query sequence and the reference sequence respectively.
- H can only be present as the first and/or last operation.
- S may only have H operations between them and the ends of the CIGAR string.
- For mRNA-to-genome alignment, an N operation represents an intron. For other types of alignments, the interpretation of N is not defined.
- Sum of lengths of the M/I/S/=/X operations shall equal the length of SEQ.
RNEXT: reference sequence name of the primary alignment of the next read in the template.
For the last read, the next read is the first read in the template. If @SQ header lines are present, RNEXT (if not ‘’ or ‘=’) must be present in one of the SQ-SN tag. This field is set as ‘’ when the information is unavailable, and set as ‘=’ if RNEXT is identical RNAME. If not ‘=’ and the next read in the template has one primary mapping (see also bit 0x100 in FLAG), this field is identical to RNAME at the primary line of the next read. If RNEXT is ‘*’, no assumptions can be made on PNEXT and bit 0x20.
PNEXT: 1-based position of the primary alignment of the next read in the template.
This field equals POS at the primary alignment of the next read. If PNEXT is 0, no assumptions can be made on RNEXT and bit 0x20. Set as 0 when the information is unavailable.
TLEN: The signed observed Template length.
If all segments are mapped to the same reference, the unsigned observed template length equals the number of bases from the leftmost mapped base to the rightmost mapped base. The leftmost segment has a plus sign and the rightmost has a minus sign. The sign of segments in the middle is undefined. It is set as 0 for single-segment template or when the information is unavailable.
SEQ: segment sequence
The sequence that was aligned. If hard clipping occurred, only the aligned portion is represented, if soft clipping occurred, the original sequence is present.
QUAL: segment quality scores
The quality scores of the sequence that was aligned. If hard clipping occurred, only the aligned portion is represented, if soft clipping occurred, the original sequence is present.
Variant descriptive file Types
Variant Call Format (VCF)
VCF (variant call format) is the standard format for variant reporting, is a text file format (possibly likely stored in a compressed manner, BCF). For an up to date scription of the specifications see VCF specifications.
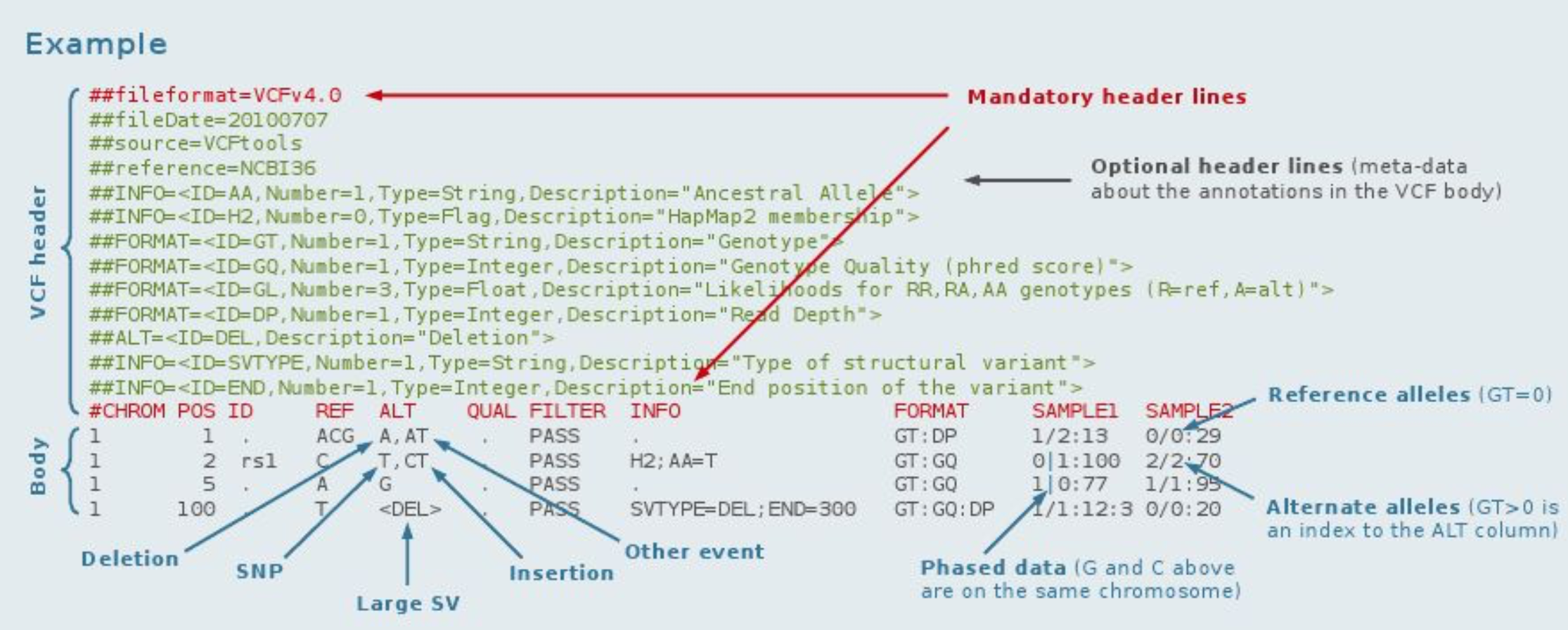
It contains meta-information lines (prefixed with “##”), a header line (prefixed with “#”), and data lines each containing information about a position in the genome and genotype information on samples for each position (text fields separated by tabs). There are 8 fixed columns followed by a column for each sample. Samples are defined by their RG tag added during the mapping process. Zero length fields ar enot allowed, a dot (“.”) must be used instead.
Meta-information Lines
File meta-information is included after the ## string and must be key=value pairs. Meta-information lines are optional, and require an ID which must be unique within their type. For all of the structured lines (##INFO, ##FORMAT, ##FILTER, etc.), extra fields can be included after the default fields. For example:
##INFO=<ID=ID,Number=number,Type=type,Description=”description”,Source=”description”,Version=”128”>
Data Lines (Columns 1 to 8)
CHROM — chromosome
An identifier from the reference genome or an angle-bracketed ID String
(“
POS — position The reference position, with the 1st base having position 1.
ID — identifier Semi-colon separated list of unique identifiers where available. If this is a dbSNP variant the rs number(s) should be used. If there is no identifier available, then the MISSING value should be used.
REF — reference base(s) Each base must be one of A,C,G,T,N (case insensitive). Multiple bases are permitted. The value in the POS field refers to the position of the first base in the String. For simple insertions and deletions in which either the REF or one of the ALT alleles would otherwise be null/empty.
ALT — alternate base(s) Comma separated list of alternate non-reference alleles. These alleles do not have to be called in any of the samples.
QUAL — quality Phred-scaled quality score for the assertion made in ALT. i.e. −10log10 prob(call in ALT is wrong). If ALT is ‘.’ (no variant) then this is −10log10 prob(variant), and if ALT is not ‘.’ this is −10log10 prob(no variant). If unknown, the MISSING value must be specified.
FILTER — filter status PASS if this position has passed all filters, i.e. a call is made at this position. Otherwise, if the site has not passed all filters, a semicolon-separated list of codes for filters that fail. e.g. “q10;s50” might indicate that at this site the quality is below 10 and the number of samples with data is below 50% of the total number of samples. ‘0’ is reserved and must not be used as a filter String. If filters have not been applied, then this field must be set to the MISSING value.
INFO — additional information INFO fields are encoded as a semicolon-separated series of short keys with optional values in the format: key[=data[,data]]. The exact format of each INFO key should be specified in the meta-information. There are several common, reserved keywords that are standards across the community. See their detailed definitions below.
- AA - Ancestral allele
- AC - Allele count in genotypes, for each ALT allele, in the same order as listed
- AD - Total read depth for each allele
- ADF - Read depth for each allele on the forward strand
- ADR - Read depth for each allele on the reverse strand
- AF - Allele frequency for each ALT allele in the same order as listed (estimated from primary data, not called genotypes)
- AN - Total number of alleles in called genotypes
- BQ - RMS base quality
- CIGAR - Cigar string describing how to align an alternate allele to the reference allele
- DB - dbSNP membership
- DP - Combined depth across samples
- END - End position (for use with symbolic alleles)
- H2 - HapMap2 membership
- H3 - HapMap3 membership
- MQ - RMS mapping quality
- MQ0 - Number of MAPQ == 0 reads
- NS - Number of samples with data
- SB - Strand bias
- SOMATIC - Somatic mutation (for cancer genomics)
- VALIDATED - Validated by follow-up experiment
- 1000G - 1000 Genomes membership
INFO keys used for structural variants
This key can be derived from the REF/ALT fields but is useful for filtering.
- DEL - Deletion relative to the reference
- INS - Insertion of novel sequence relative to the reference
- DUP - Region of elevated copy number relative to the reference
- INV - Inversion of reference sequence
- CNV - Copy number variable region (may be both deletion and duplication)
- BND - Breakend
Genotype fields (Columns 9 and above)
If genotype information is present, then the same types of data must be present for all samples. First a FORMAT field (Column 9) is given specifying the data types and order (colon-separated FORMAT keys from the meta-information section). This is followed by one data block per sample, with the colon- separated data corresponding to the types specified in the format. The first key must always be the genotype (GT) if it is present. If any of the fields is missing, it is replaced with the MISSING value. As with the INFO field, there are several common, reserved keywords that are standards across the community. See their detailed definitions below.
- AD - Read depth for each allele
- ADF - Read depth for each allele on the forward strand
- ADR - Read depth for each allele on the reverse strand
- DP - Read depth
- EC - Expected alternate allele counts
- FT - Filter indicating if this genotype was “called”
- GL - Genotype likelihoods
- GP - Genotype posterior probabilities
- GQ - Conditional genotype quality
- GT - Genotype
- HQ - Haplotype quality
- MQ - RMS mapping quality
- PL - Phred-scaled genotype likelihoods rounded to the closest integer
- PQ - Phasing quality
- PS - Phase set
Variant effect Prediction ANN field
- VCF INFO field name ANN, stands for ‘annotations’
- Data fields are encoded separated by pipe sign “|”; the order of fields is written in the VCF header.
- When comparing genomic coordinates, the comparison should be done first by chromosome names (compared alphabetically), then by start position, and finally by end position.
-
Special characters: Comma, space, tab, newline or pipe characters (‘,’, ‘ ‘, ‘\t’, ‘\n’, ‘ ’, etc.) can be converted to underscore (‘_’) (This is the preferred way). - Multiple “effects / consequences” are separated by comma.
See the current standard for a more thorough description.