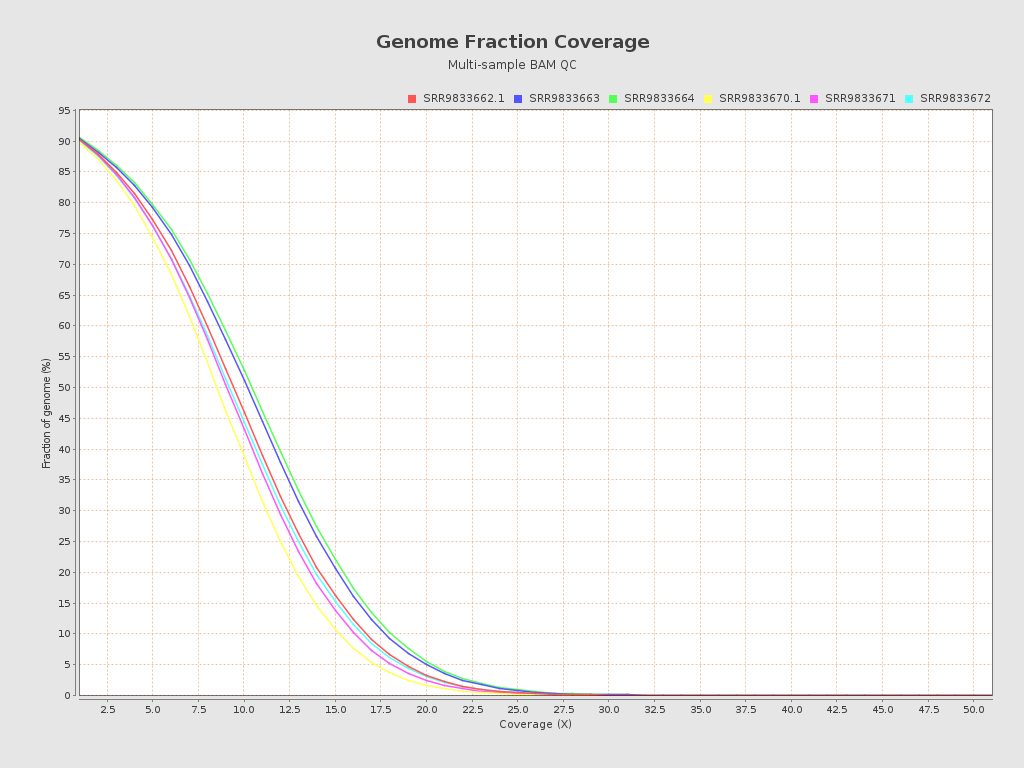
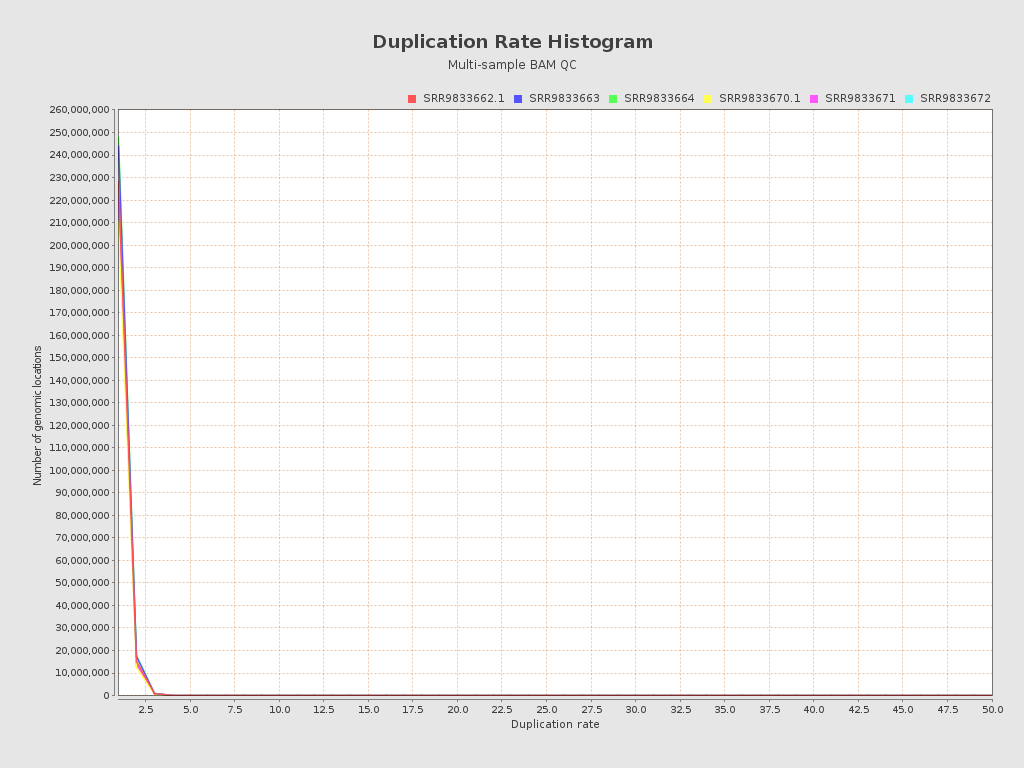
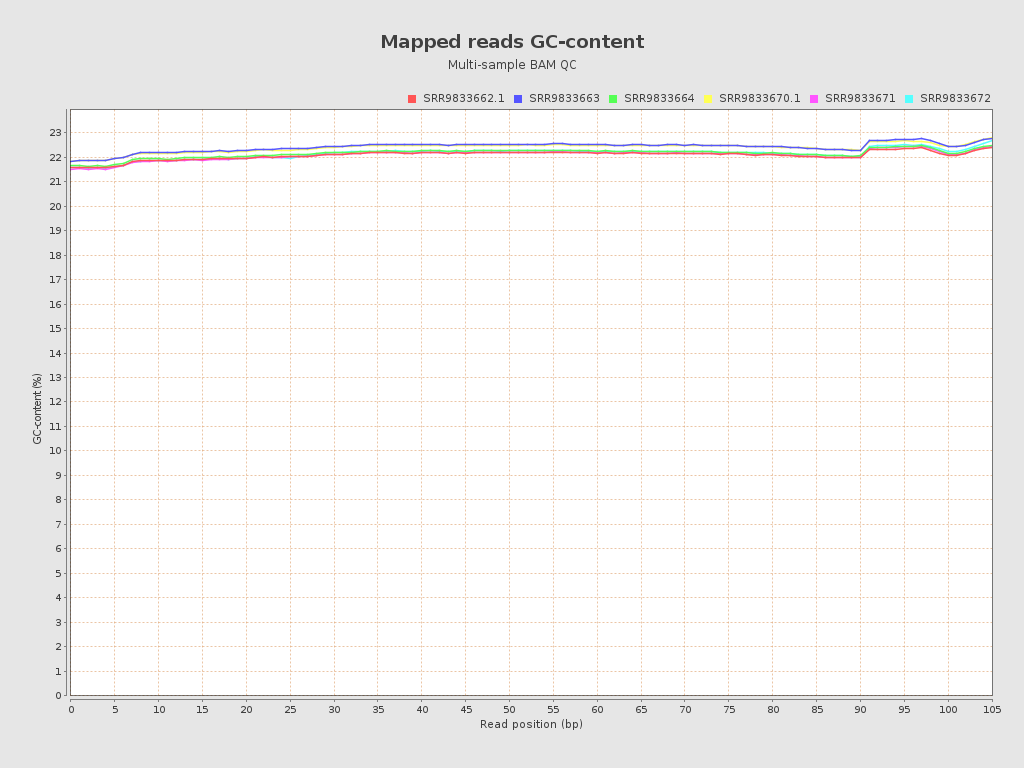
Input data and parameters
Samples
| SRR9833663 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833663 |
| SRR9833662.1 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833662.1 |
| SRR9833664 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833664 |
| SRR9833670.1 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833670.1 |
| SRR9833671 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833671 |
| SRR9833672 | /share/biocore/projects/Internal_Jessie_UCD/Workshops/Epigenetics/04-Qualimap/SRR9833672 |
Summary
Globals
| Number of samples | 6 |
| Total number of mapped reads | 1,587,209,058 |
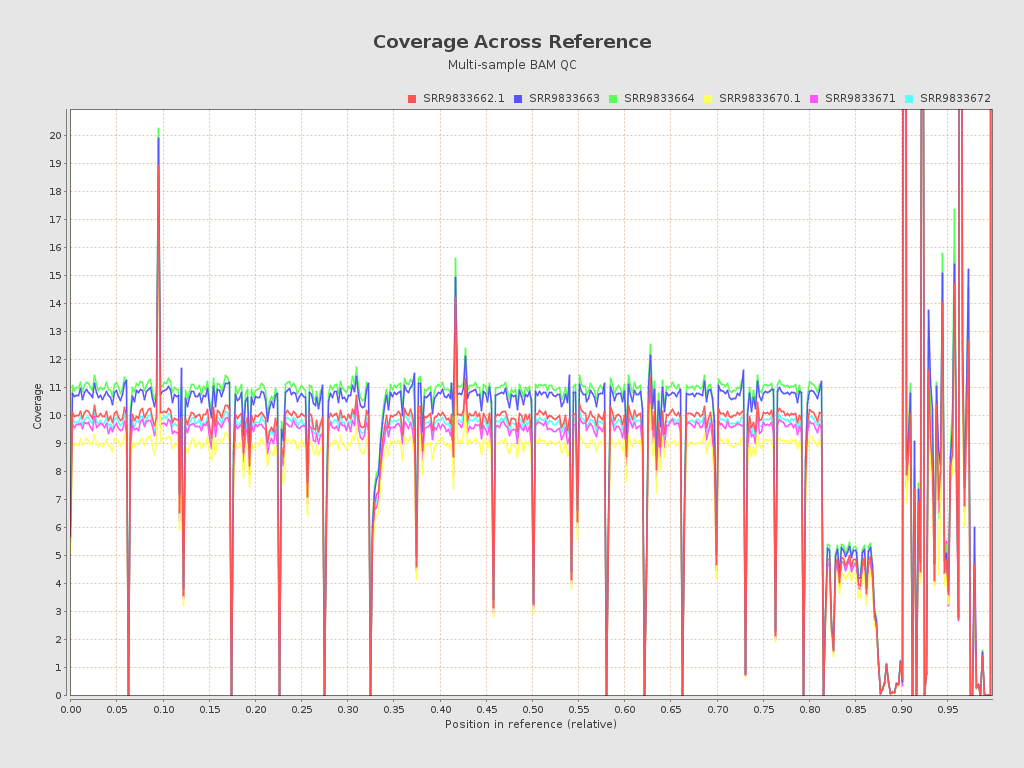
| Mean samples coverage | 9.08 |
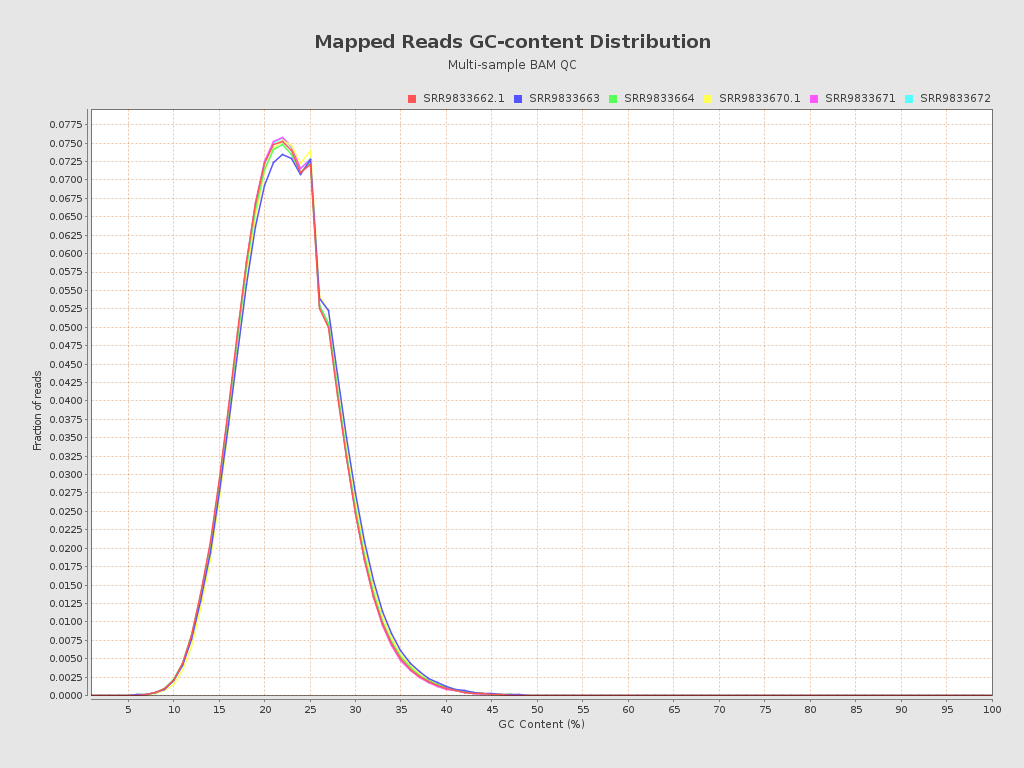
| Mean samples GC-content | 22.2 |
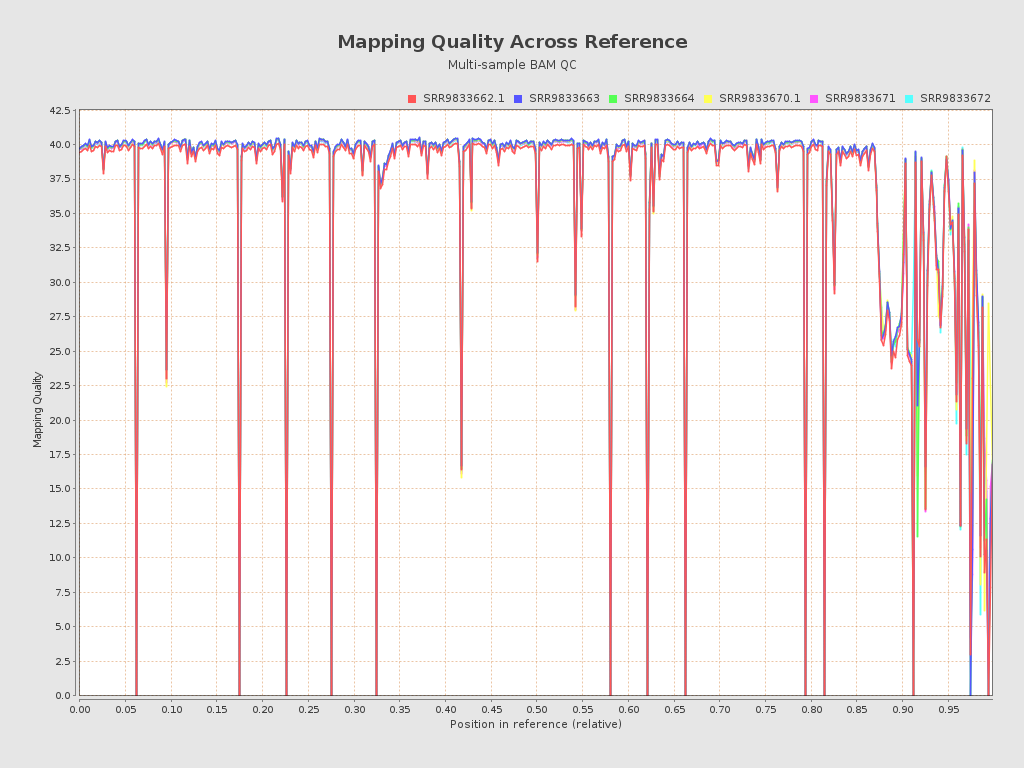
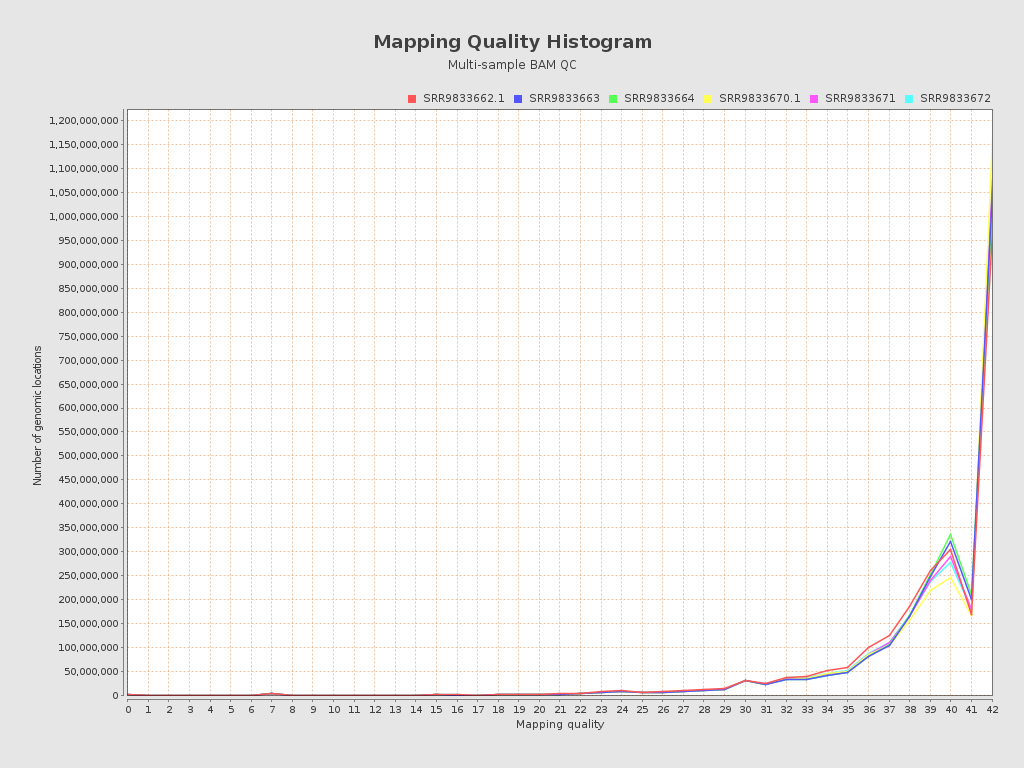
| Mean samples mapping quality | 37.15 |
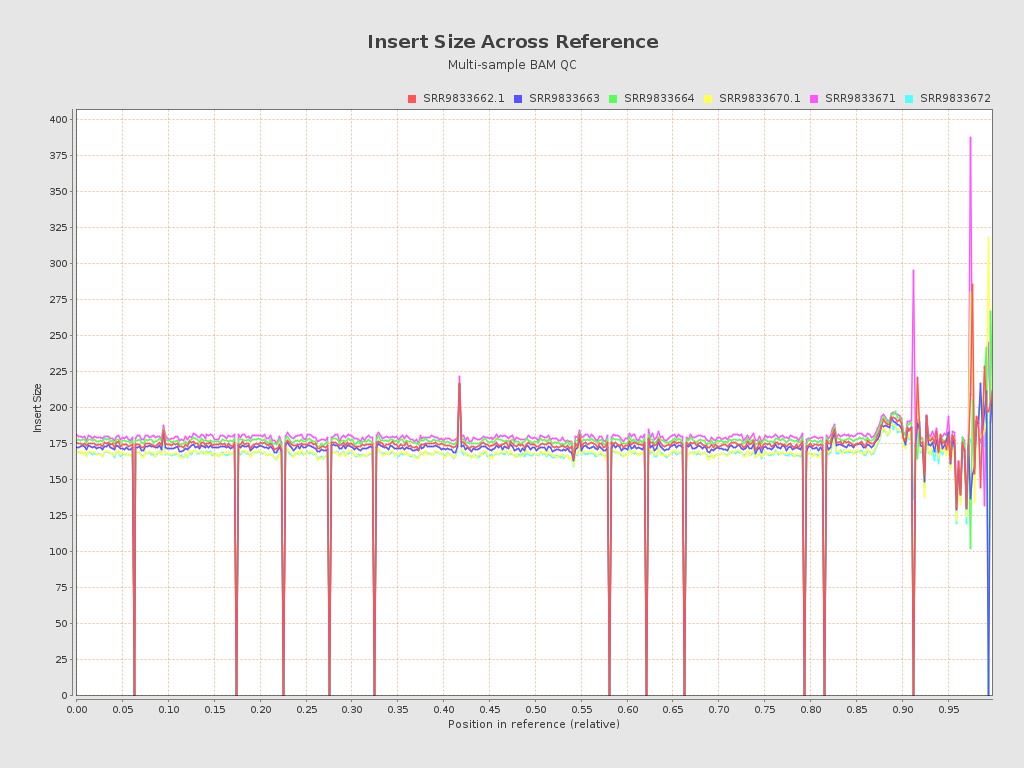
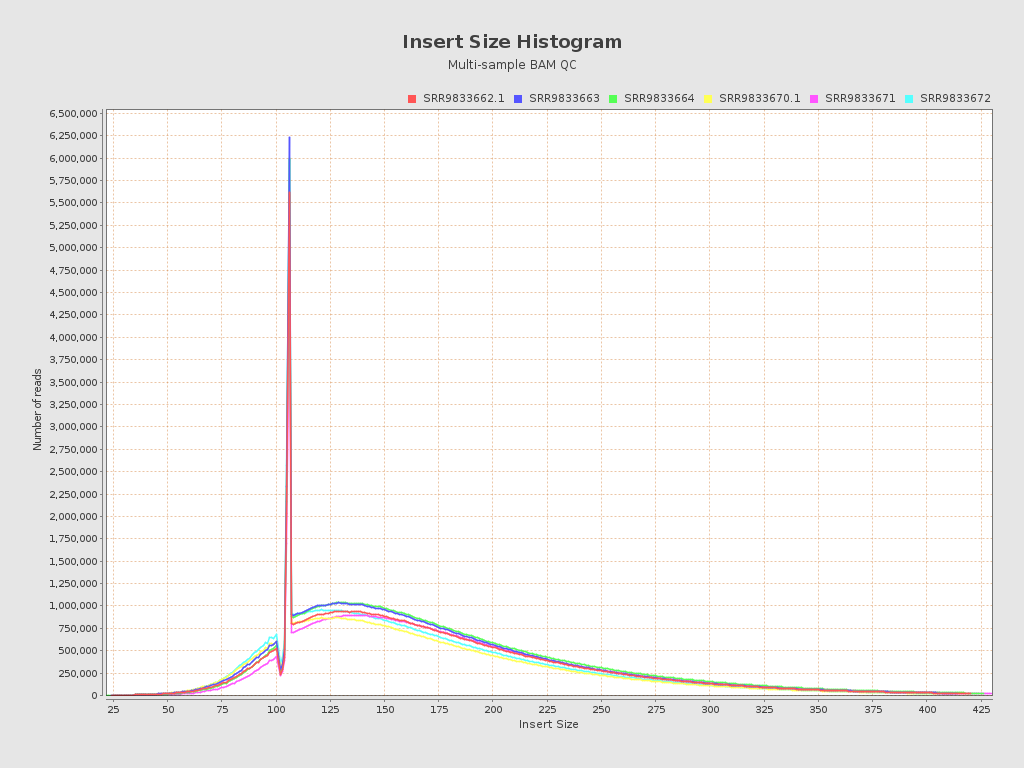
| Mean samples insert size | 157.5 |
Sample statistics
| Sample name | Coverage mean | Coverage std | GC percentage | Mapping quality mean | Insert size median |
| SRR9833662.1 | 9.0506 | 39.8298 | 22.07 | 36.8654 | 159.0 |
| SRR9833663 | 9.7366 | 40.8321 | 22.4 | 37.2052 | 157.0 |
| SRR9833664 | 9.9595 | 41.5974 | 22.15 | 37.1786 | 161.0 |
| SRR9833670.1 | 8.1623 | 39.3772 | 22.38 | 37.2802 | 152.0 |
| SRR9833671 | 8.6892 | 39.7925 | 22.07 | 37.1512 | 164.0 |
| SRR9833672 | 8.8532 | 39.7 | 22.12 | 37.2193 | 152.0 |

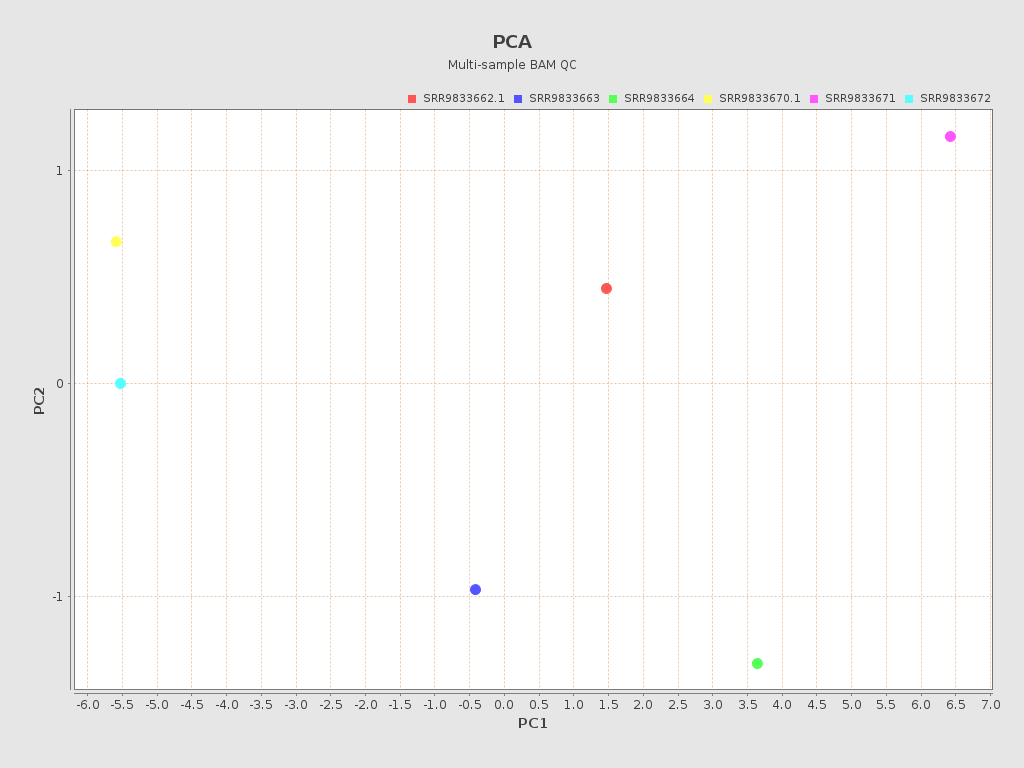
.png)