Closing Thoughts
The forest through the trees.
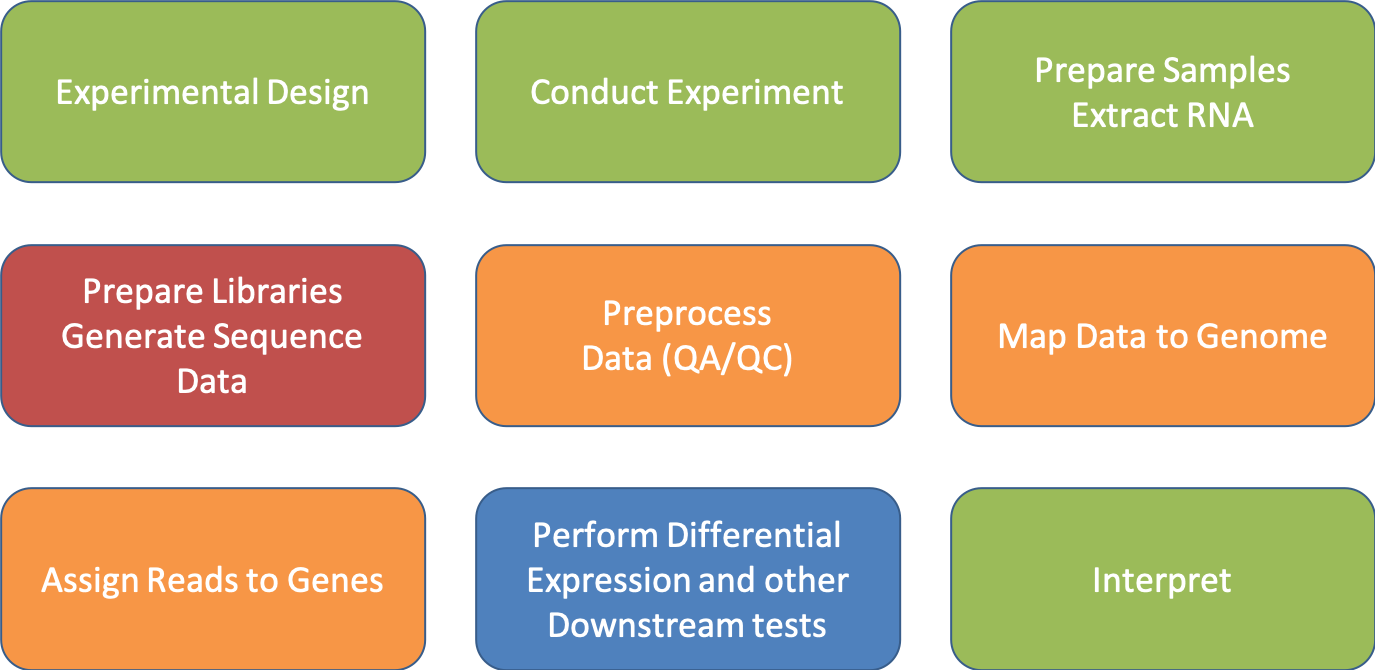
Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
Prerequisites for bioinformatics
- Access to a multi-core (24 cpu or greater), ‘high’ memory 64Gb or greater Linux server.
- Familiarity with the ’command line’ and at least one programming language.
- Basic knowledge of how to install software
- Basic knowledge of R (or equivalent) and statistical programming
- Basic knowledge of Statistics and model building
The bottom line
- Spend the time (and money) planning and producing good quality, accurate and sufficient data for your experiment.
- Get to know to your data, develop and test expectations
- Result, you’ll spend much less time (and less money) extracting biological significance and results during analysis.
Workshop Week 2 reservation
workshop ACTIVE 2019-03-09T10:46:16 2019-03-22T00:00:00 12-12:13:44 drove-[4,13,18]
My recommendation is to follow all of the instructions again, from the beginning on your own and send emails to
bioinformatics.training@ucdavis.edu
And we will be responsive to answering questions
Upcoming Workshops
Now in our 12th year of training researchers, the UC Davis Bioinformatics Core invites you to attend one or more of our 2019 workshops. The following workshops are suitable for beginners, and are also of interest to those with some data analysis experience.
For more information on these and other events, and to register, go to https://registration.genomecenter.ucdavis.edu/
March 11-14: RNA-Seq Workshop @ UCSF
August 19-23: RNA-Seq Workshop @ UC Davis
This popular workshop contains a rich collection of lectures and hands-on sessions, covering both theory and tools associated with command-line RNA-seq data analysis. Topics include experimental design, cost estimation, data generation, and analysis of RNA-Seq data generated on the Illumina sequencing platform, as well as post differential expression analyses, such as gene set enrichment analysis, Gene Ontology/Pathway analysis, and generation of relevant figures and tables.
Participants will explore software and protocols, create and modify workflows, and diagnose/treat problematic data utilizing high performance computing services. Exercises will be performed with provided datasets, using command-line interaction and with RStudio. (This workshop does not cover single-cell RNA-Seq)
For more information and to register, go to https://registration.genomecenter.ucdavis.edu/.
June 24-27: Single-Cell RNA-Seq Workshop @ UC Davis
July 1-3: Single-Cell RNA-Seq Workshop @ UCSF
This workshop will cover experimental design, data generation, and analysis of single-cell RNA sequencing data (primarily generated using the 10x platform) on the command line and within the R statistical programming language. Participants will explore software and protocols, create and modify workflows, and diagnose/treat problematic data utilizing high performance computing services. The primary packages used for analysis will be 10x software (for sequence reads to counts) and the R packages (e.g, Seurat) for downstream analysis.
For more information and to register, go to https://registration.genomecenter.ucdavis.edu/.
September 3-6: Microbial Community Analysis @ UC Davis
This workshop covers amplicon-based microbial analysis and methodology using high throughput sequencing technologies. Topics will include:
- Differences between communty analysis with amplicons versus shotgun metagenomics and metatranscriptomics
- Experimental design
- Sequence data quality assurance and expectations
- Processing of amplicon reads (using the python application dbcAmplicons)
- Analysis of community structure (using R and vegan)
- Lecture on metagenome assembly and metatranscriptome assembly and annotation
- Post analysis and visualization (using R)
Exercises will be performed with provided datasets, using command-line interaction and with RStudio.
For more information and to register, go to https://registration.genomecenter.ucdavis.edu/.
December: Bioinformatics Prerequisites Workshop @ UC Davis
This workshop will cover the basic prerequisites required before performing bioinformatics analysis, including basic command line and high performance computing concepts, how to install software, use help, run applications, and advanced command line (shell scripting, pipes, etc.). Also covered will be basic R programming, basic statistical concepts, and model building. There are no prerequisites for this workshop other than an interest in bioinformatics!
If you would like to take a look at the documentation from previous workshops, see https://bioinformatics.ucdavis.edu/training/documentation/.
Questions? Contact us at training.bioinformatics@ucdavis.edu Email List Note: If you received this email directly from training.bioinformatics@ucdavis.edu, you are currently subscribed to one of our announcements lists. To subscribe / unsubscribe from the Bioinformatics Core email lists, please follow the instructions at the bottom of http://bioinformatics.ucdavis.edu/contact-us/.