Annotation via biomart
Annotation from BioMart with Ensembl names is most flexible way to retrieve tabular annotation for an organism.
1. The Biomart start page should look like …
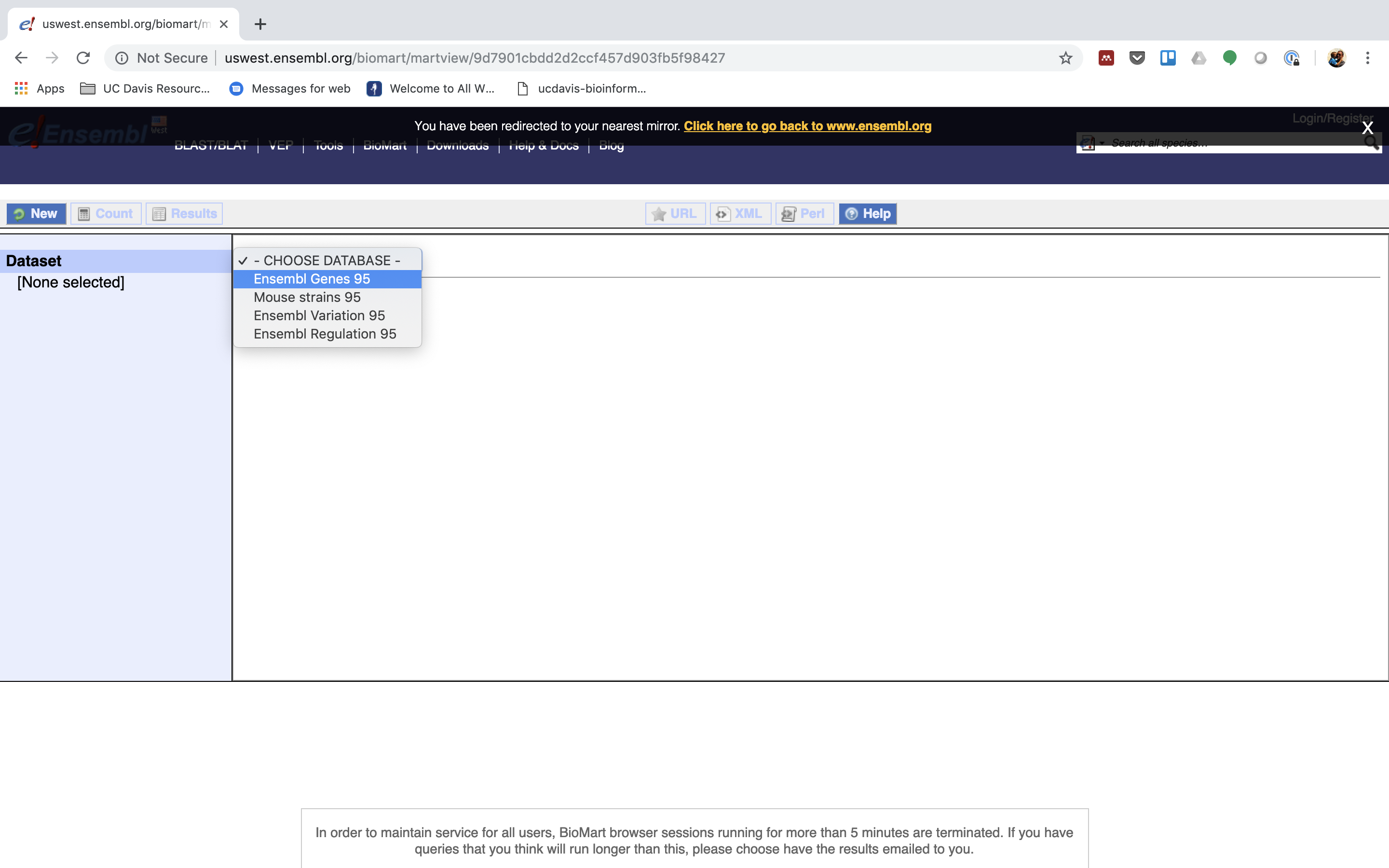
2. First select the dataset, for gene expression experiment select Ensembl Genes 95 (version 95).
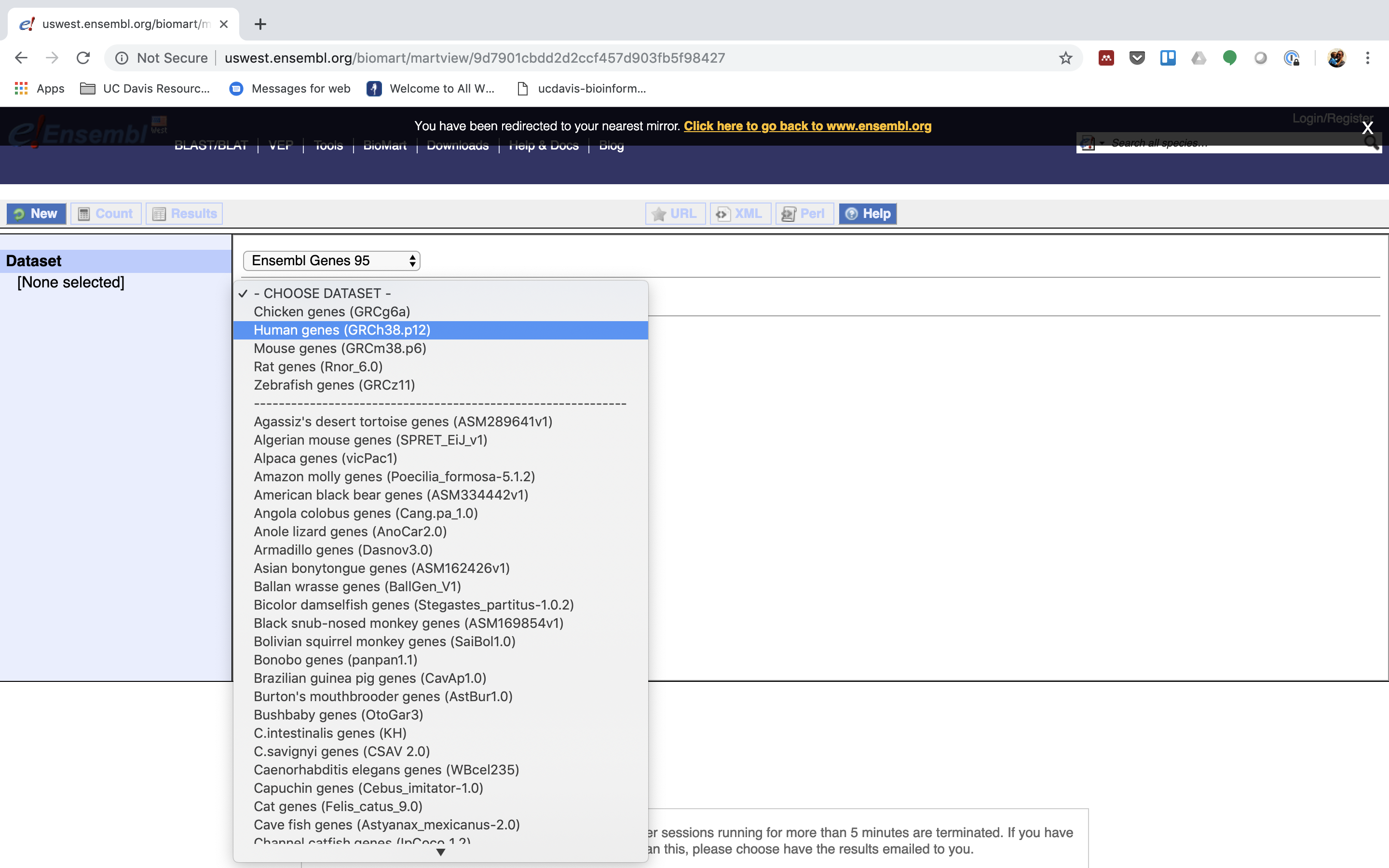
3. Then the Organism, Here Human genes which is based on the GRC38.p12 genome.
4. You can choose to filter to only a subset of genes. Or a chromosome, or regions. We won’t filter here. BY default, all genes in the genome are selected.
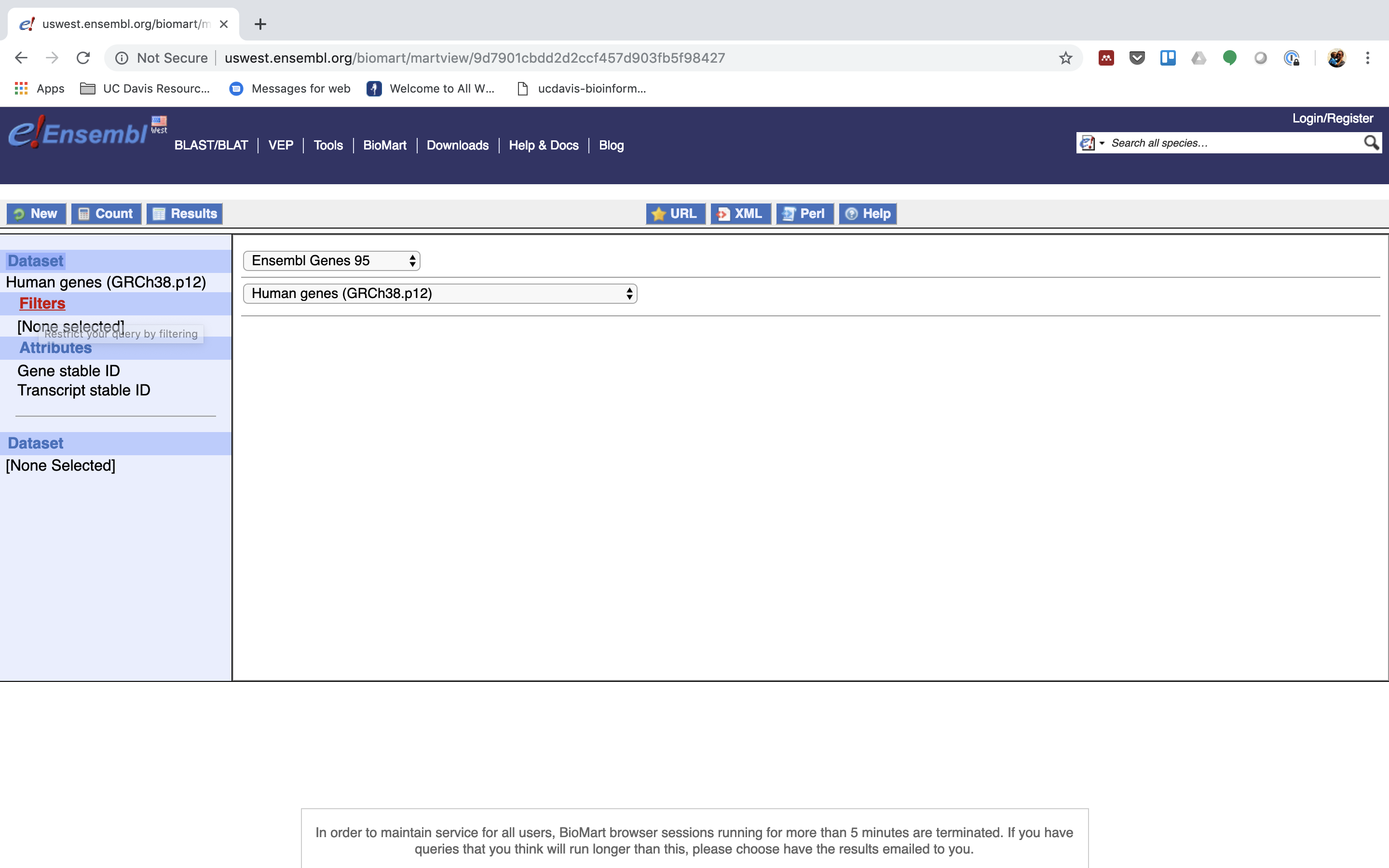
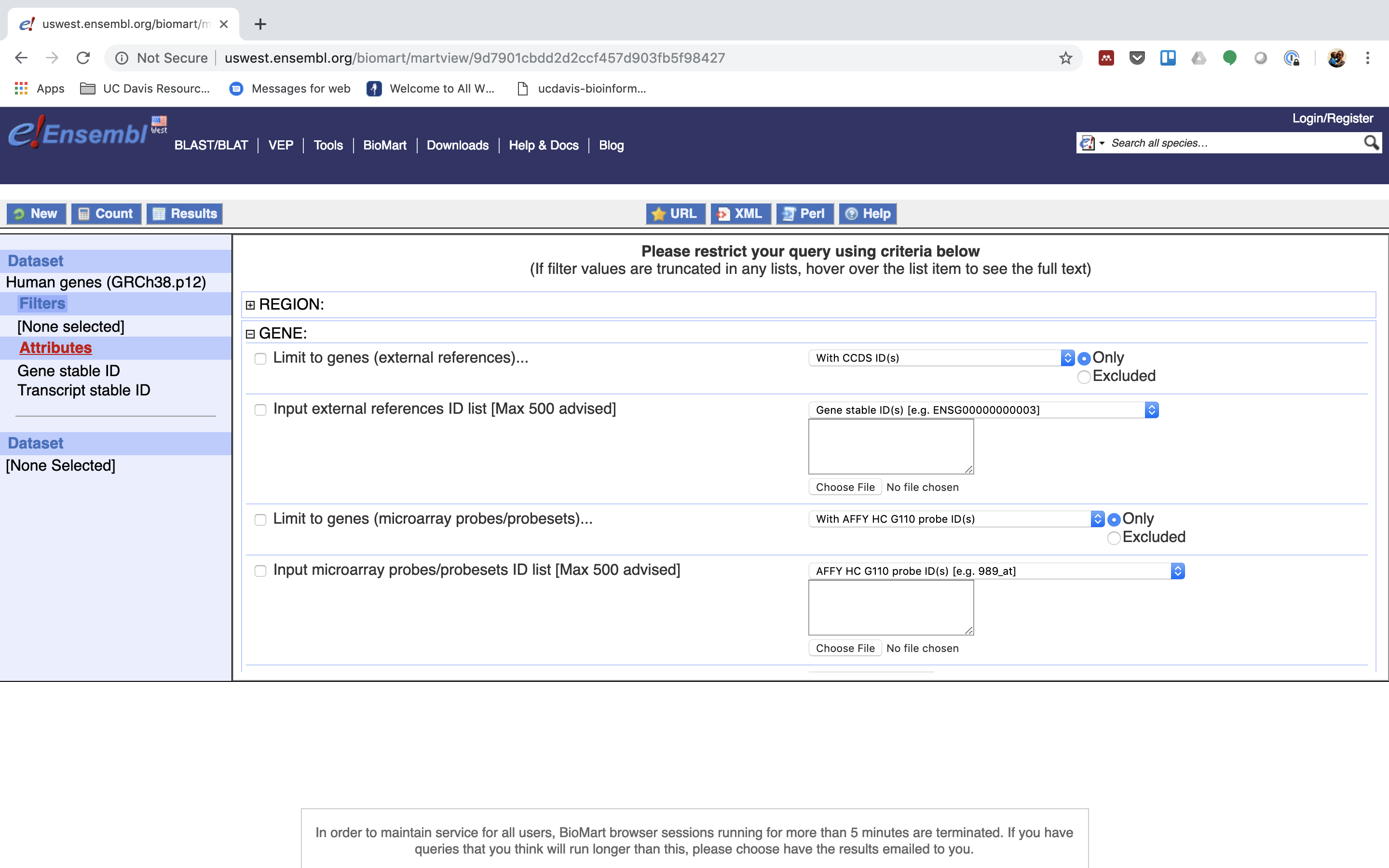
5. Next select the attributes you want in the table.
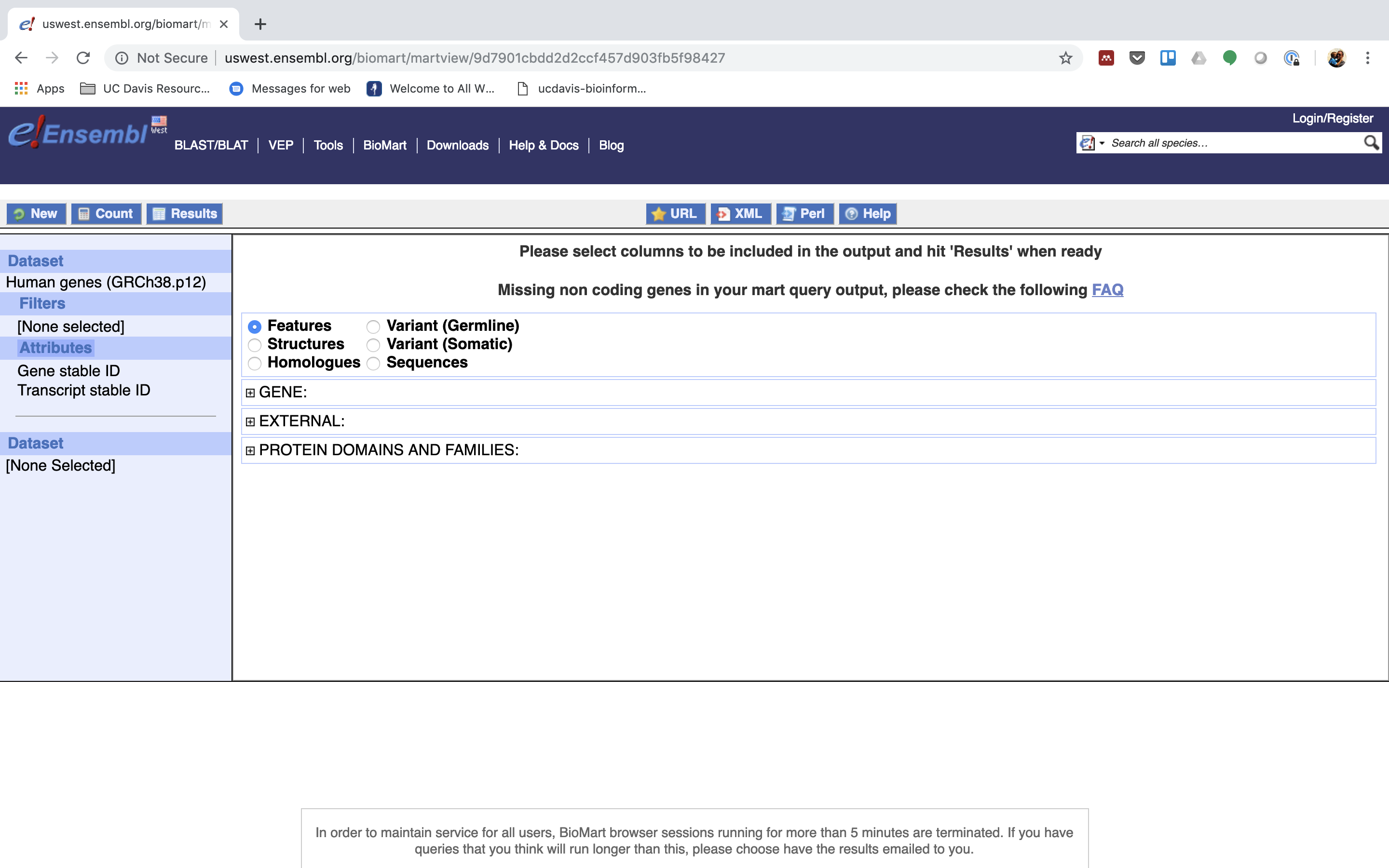
6. Expand the ‘GENE’ tab.
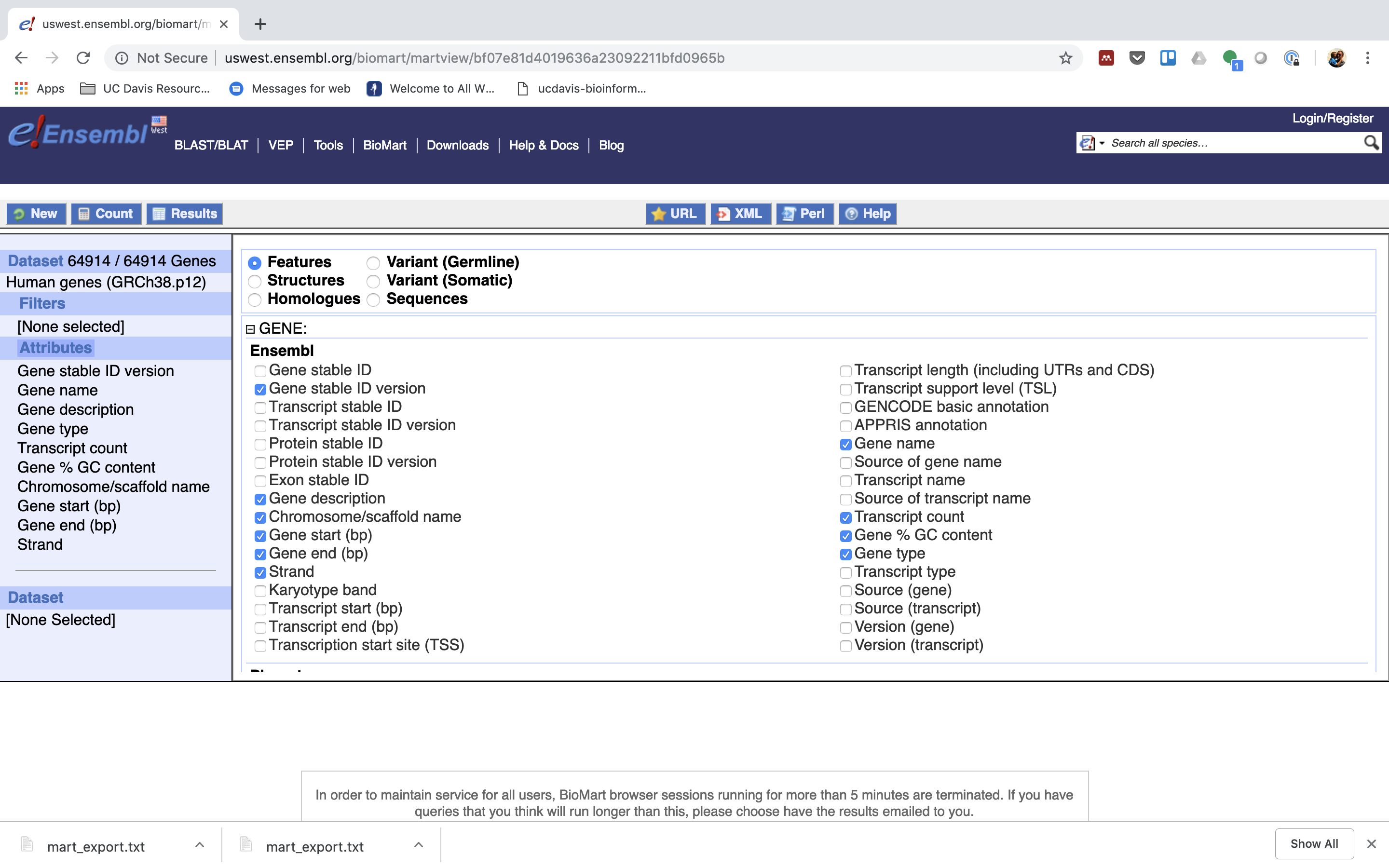
7. Click “Results” (Top left -ish).
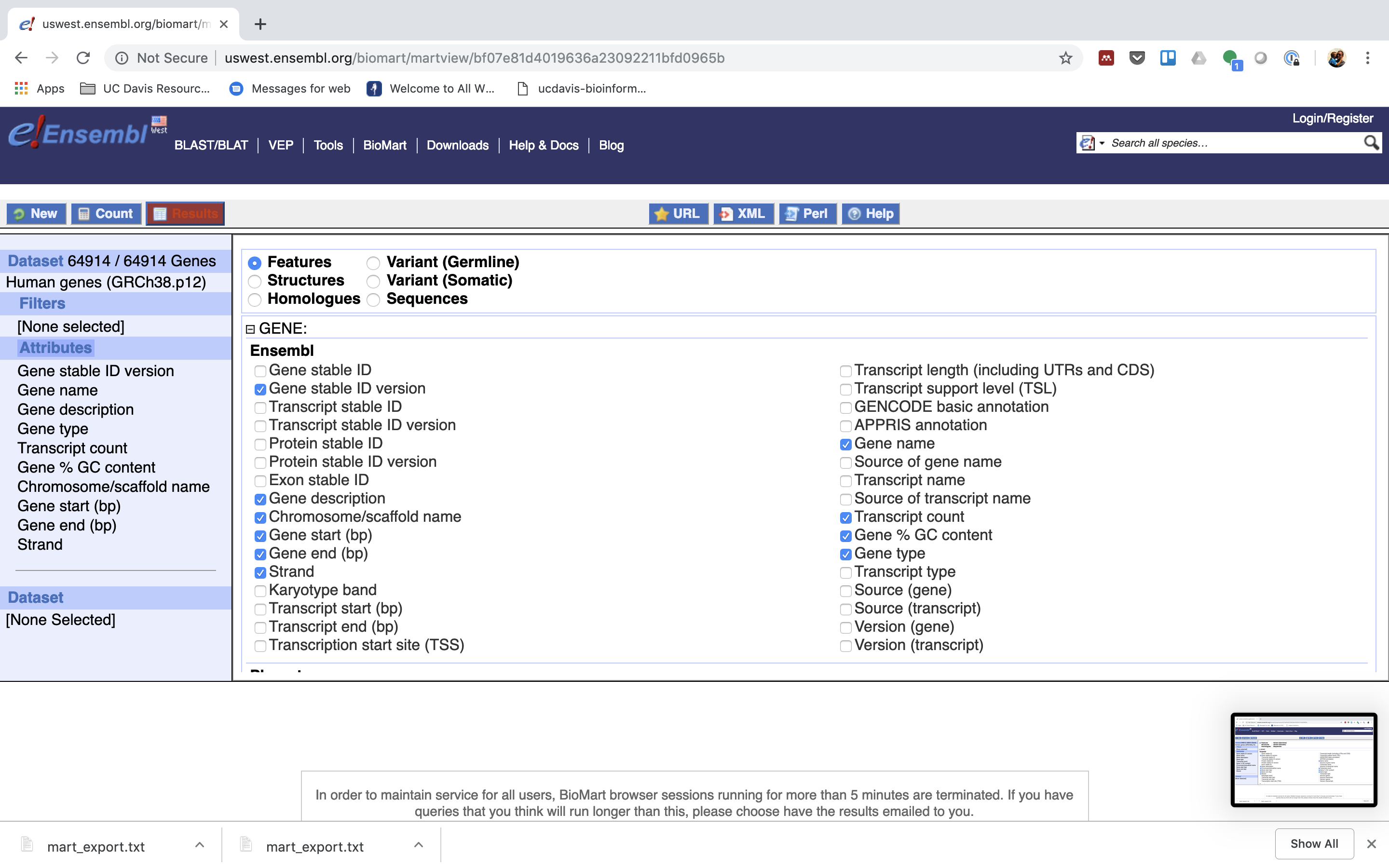
7. Select “GO”, to download a tab-separated value (tsv) file.
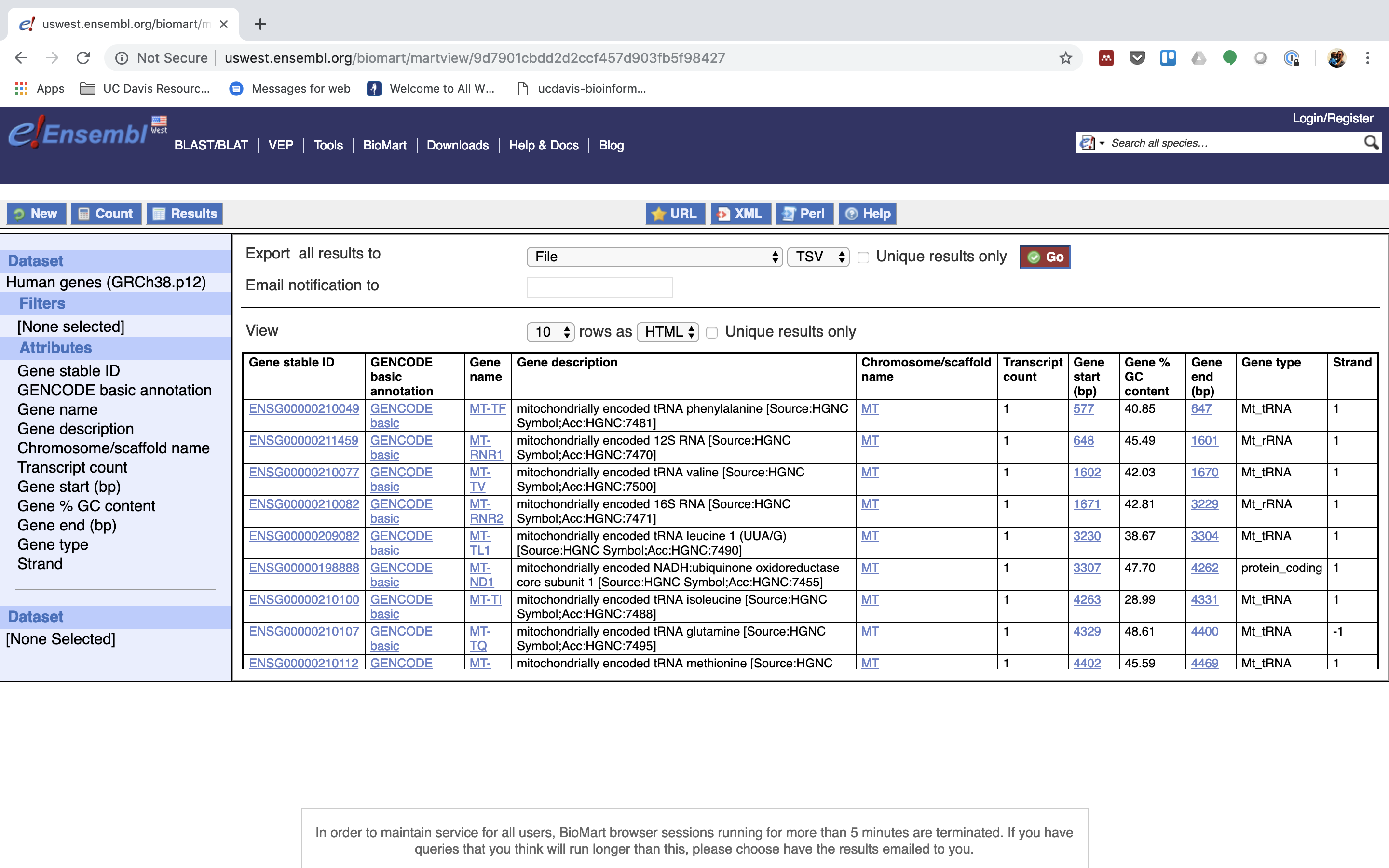
8. Put the file into our working directory, rename to “ensembl_hg_95.tsv” and open the file in Excel to view the annotation.