Pairwise correlations of normalized counts on same sample
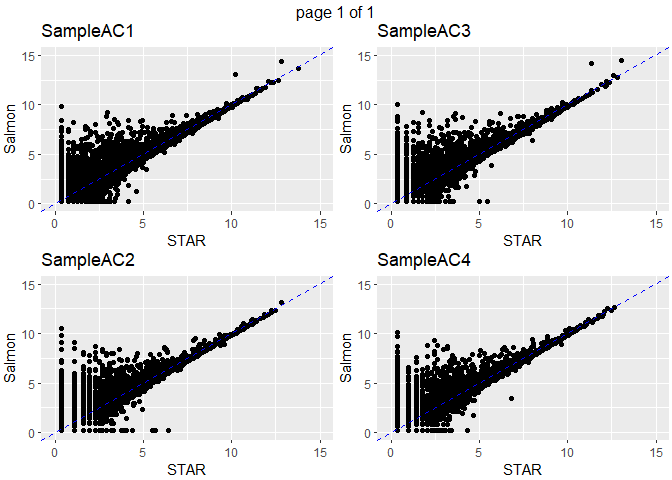
(Figure shows 4 samples, these are representative of pattern on remaining samples.)
Note the pattern of genes with low expression with STAR and high expression with Salmon.
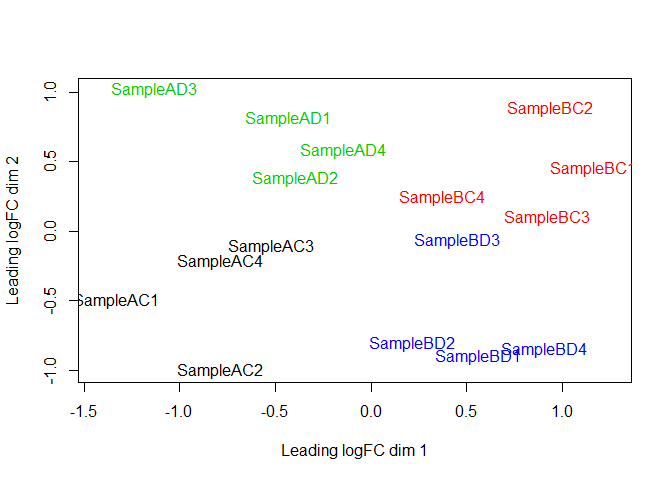
MDS plot, STAR counts:
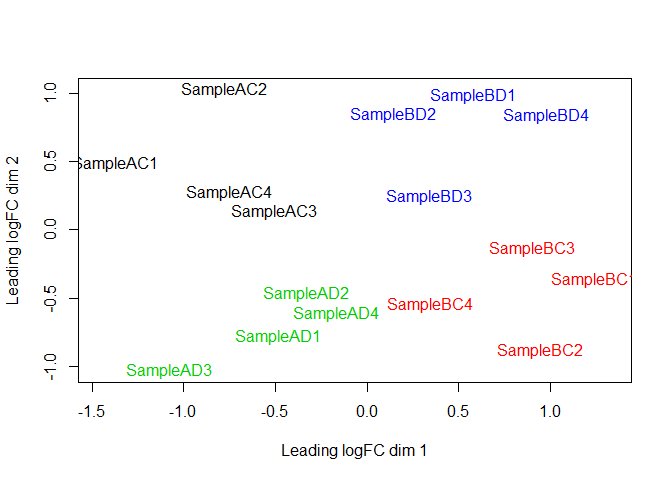
MDS plot, Salmon counts
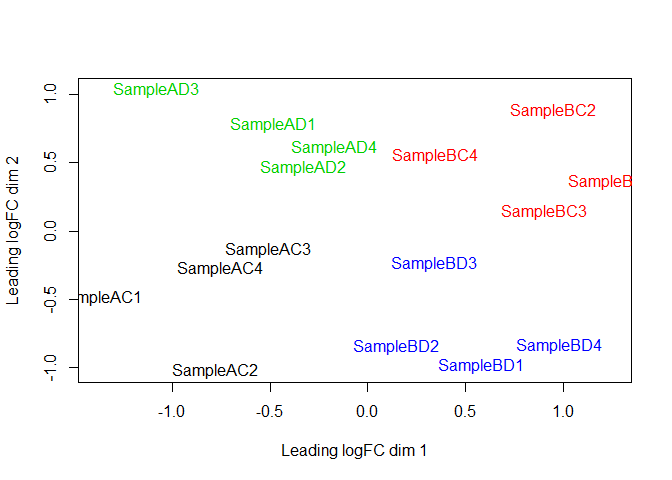
Note that you can rotate or flip an MDS plot without changing the distances between samples (the orientation is arbitrary). Here’s the Salmon MDS plot rotated to match the orientation of the STAR MDS plot:
Top 10 genes with STAR
## Gene.stable.ID HGNC.symbol logFC AveExpr t P.Value
## 1 ENSG00000129824 RPS4Y1 -6.014740 3.826209 -7.151711 7.192005e-07
## 2 ENSG00000067048 DDX3Y -6.403500 2.913309 -6.830168 1.382919e-06
## 3 ENSG00000012817 KDM5D -6.390275 2.940730 -6.718986 1.739325e-06
## 4 ENSG00000198692 EIF1AY -5.184644 1.887900 -6.693343 1.834216e-06
## 5 ENSG00000099725 PRKY -5.826135 2.973758 -6.443233 3.093471e-06
## 6 ENSG00000183878 UTY -5.060957 2.437612 -6.356324 3.716707e-06
## 7 ENSG00000111052 LIN7A -1.675545 5.558748 -6.134283 5.966643e-06
## 8 ENSG00000114374 USP9Y -5.196104 1.973817 -5.754669 1.359469e-05
## 9 ENSG00000131002 TXLNGY -5.396197 2.403759 -5.741837 1.398266e-05
## 10 ENSG00000173597 SULT1B1 -1.348515 6.533206 -5.420129 2.848455e-05
## adj.P.Val B
## 1 0.006643531 4.687789
## 2 0.006643531 2.965094
## 3 0.006643531 2.974988
## 4 0.006643531 2.138306
## 5 0.008963642 3.051457
## 6 0.008974608 2.509162
## 7 0.012349247 4.011984
## 8 0.022508972 1.508833
## 9 0.022508972 1.847063
## 10 0.041268410 2.625203
Top 10 genes with Salmon
## Gene.stable.ID HGNC.symbol logFC AveExpr t P.Value
## 1 ENSG00000129824 RPS4Y1 -5.964371 3.665746 -6.954325 1.386850e-06
## 2 ENSG00000067048 DDX3Y -6.352257 2.759204 -6.775343 1.978005e-06
## 3 ENSG00000198692 EIF1AY -4.964644 1.794527 -6.527168 3.259416e-06
## 4 ENSG00000012817 KDM5D -6.294545 2.770694 -6.495961 3.472717e-06
## 5 ENSG00000099725 PRKY -4.659910 3.160600 -6.220045 6.116606e-06
## 6 ENSG00000183878 UTY -4.913783 2.241138 -6.133448 7.320855e-06
## 7 ENSG00000111052 LIN7A -1.670540 5.402575 -5.983866 1.000851e-05
## 8 ENSG00000131002 TXLNGY -5.260164 2.217746 -5.598758 2.267707e-05
## 9 ENSG00000114374 USP9Y -5.072640 1.879345 -5.494636 2.837586e-05
## 10 ENSG00000131724 IL13RA1 -1.155352 7.144644 -5.286023 4.462809e-05
## adj.P.Val B
## 1 0.01257210 4.142661
## 2 0.01257210 2.827628
## 3 0.01257210 2.044915
## 4 0.01257210 2.619452
## 5 0.01766888 3.199943
## 6 0.01766888 2.143609
## 7 0.02070475 3.546144
## 8 0.04104833 1.561104
## 9 0.04565676 1.157373
## 10 0.06462593 2.223904
10 genes are DE (adjusted P < 0.05) with STAR, and 9 genes are DE with Salmon. All genes DE with Salmon are DE with STAR.
Conclusions
- STAR and Salmon show differences in quantification of low expressed genes.
- This does not seem to have much impact on the relative distances between samples or differential expression (note that low expressed genes are often filtered out before analysis).
- Pick one approach for a project and go with it.
- Don’t run multiple analyses and pick the one that gives you the most DE genes.
