Hands-on Workshop Assembling and Annotating Bacterial Genomes Using PacBio Long Reads
This workshop aims to provide to the participants the basic knowledge from beginning to end to assemble near-complete or complete high-quality bacterial genomes from PacBio long reads.
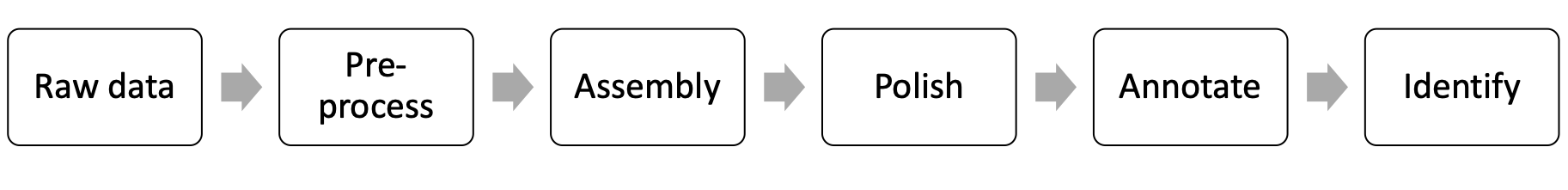
- Introduction to PacBio sequencing data. Matthew Settles; Univ. of California, Davis, Davis, CA
- Bacterial genome assembling with PacBio data. Hannah Lyman; Univeristy of California, Davis, Davis, CA
- Annotating High-quality Bacterial Genomes. Jie Li; Univ. of California, Davis, Davis, CA
- Bacterial Taxonomy with Whole-genome Data. Maria Bonatelli; Univ. of São Paulo, Piracicaba, Brazil
The bacterium Bacillus thuringiensis RZ2MS9
Today, you will assemble the genome of a bacterium that was isolated from the Amazon rainforest, in the city of Maués, Amazonas state, Brazil. This bacterium was isolated from the rizosphere of guarana plants (Paullinia cupana).

The Bacillus thuringiensis RZ2MS9 is a plant growth-promoting bacteria, so it harbors several genes related with plant growth-promoting traits, such as the production of indole acetic acid, solubilization of phosphate, and more.

Read the paper: Batista et al. 2018.
Computing needs
The quantity of data storage and the amount of processing needed should be sufficient on most laptop systems; however, many of the application used in the workshop requires the ability to compile code on a command line. As such we expect you to have these tools available on your system in order to fully participate.
The applications that need to be first installed are:
Data Availability
The data files needed for this workshop can be found here on our Bioshare application. Please download all these data to your computers before continuing on.
Workshop Materials
-
This Bacterial Genome Assembly Workshop
https://ucdavis-bioinformatics-training.github.io/2021-ASM-genome-assembly/
