7 Introduction to the tidyverse
The tidyverse is a collection of packages by the creators of RStudio that share an approach to data science.
The authors model data science like this:
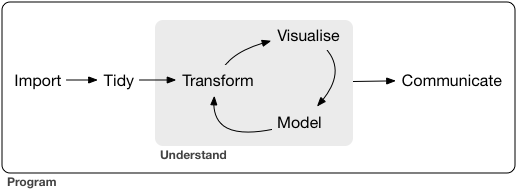
Data science life cycle
The tidyverse packages replace some of the base R functions with alternatives that are intended to be more user friendly for data scientists who are following this life cycle.
We will only be covering a few of the packages from the tidyverse.
library(tidyr)
library(dplyr)
library(magrittr)7.1 Defining tidy data
In “tidy” data, every column is a variable, every row is an observation, and every cell contains a single observation. Is the birthweight data frame tidy? Why or why not?
7.2 Pipes: combining tidyverse functions
The tidyverse employs piping to send the output of one function to another function, rather than the nesting used in base r. The “pipe” is written with a greater than symbol sandwiched between two percent signs, like this: %>%.
experiment %>%
filter(low.birthweight == TRUE) %>%
select(birth.date, length, birthweight, smoker)| birth.date | length | birthweight | smoker |
|---|---|---|---|
| 7/18/1968 | 46 | 2.05 | TRUE |
| 9/16/1968 | 48 | 1.92 | TRUE |
| 1/8/1968 | 47 | 2.66 | TRUE |
| 3/23/1967 | 50 | 2.51 | TRUE |
| 9/27/1968 | 43 | 2.65 | FALSE |
| 4/2/1968 | 48 | 2.37 | TRUE |
# equivalent to:
experiment[experiment$low.birthweight == TRUE, c("birth.date", "length", "birthweight", "smoker")]| birth.date | length | birthweight | smoker | |
|---|---|---|---|---|
| 5 | 7/18/1968 | 46 | 2.05 | TRUE |
| 8 | 9/16/1968 | 48 | 1.92 | TRUE |
| 10 | 1/8/1968 | 47 | 2.66 | TRUE |
| 13 | 3/23/1967 | 50 | 2.51 | TRUE |
| 33 | 9/27/1968 | 43 | 2.65 | FALSE |
| 35 | 4/2/1968 | 48 | 2.37 | TRUE |
7.3 Transforming data
The separate() function makes the conversion of the “birth.date” column into “month,” “day,” and “year” trivial.
experiment %>%
filter(low.birthweight == TRUE) %>%
select(birth.date, length, birthweight, smoker) %>%
separate(col = birth.date, sep = "[/]", into = c("month", "day", "year"))| month | day | year | length | birthweight | smoker |
|---|---|---|---|---|---|
| 7 | 18 | 1968 | 46 | 2.05 | TRUE |
| 9 | 16 | 1968 | 48 | 1.92 | TRUE |
| 1 | 8 | 1968 | 47 | 2.66 | TRUE |
| 3 | 23 | 1967 | 50 | 2.51 | TRUE |
| 9 | 27 | 1968 | 43 | 2.65 | FALSE |
| 4 | 2 | 1968 | 48 | 2.37 | TRUE |
The mutate() function adds a new column based on data contained in the existing columns.
experiment %>%
filter(low.birthweight == TRUE) %>%
select(birth.date, length, birthweight, smoker) %>%
mutate(d = birthweight / length)| birth.date | length | birthweight | smoker | d |
|---|---|---|---|---|
| 7/18/1968 | 46 | 2.05 | TRUE | 0.0445652 |
| 9/16/1968 | 48 | 1.92 | TRUE | 0.0400000 |
| 1/8/1968 | 47 | 2.66 | TRUE | 0.0565957 |
| 3/23/1967 | 50 | 2.51 | TRUE | 0.0502000 |
| 9/27/1968 | 43 | 2.65 | FALSE | 0.0616279 |
| 4/2/1968 | 48 | 2.37 | TRUE | 0.0493750 |
7.4 Summarizing data
The group_by() and summarize() functions apply a function to a group defined by one or more categorical variables.
experiment %>%
group_by(smoker) %>%
summarize(mean.birthweight = mean(birthweight))| smoker | mean.birthweight |
|---|---|
| FALSE | 3.509500 |
| TRUE | 3.134091 |
experiment %>%
group_by(smoker, low.birthweight) %>%
summarize(mean.birthweight = mean(birthweight))## `summarise()` has grouped output by 'smoker'. You can override using the `.groups` argument.| smoker | low.birthweight | mean.birthweight |
|---|---|---|
| FALSE | FALSE | 3.554737 |
| FALSE | TRUE | 2.650000 |
| TRUE | FALSE | 3.378824 |
| TRUE | TRUE | 2.302000 |
To change the order of rows, use arrange(). To return one or more specified rows, use slice().
experiment %>%
group_by(smoker) %>%
select(smoker, birthweight, length, head.circumference, weeks.gestation) %>%
slice_max(order_by = birthweight, n = 5)| smoker | birthweight | length | head.circumference | weeks.gestation |
|---|---|---|---|---|
| FALSE | 4.55 | 56 | 34 | 44 |
| FALSE | 4.32 | 53 | 36 | 40 |
| FALSE | 4.10 | 58 | 39 | 41 |
| FALSE | 4.07 | 53 | 38 | 44 |
| FALSE | 3.94 | 54 | 37 | 42 |
| TRUE | 4.57 | 58 | 39 | 41 |
| TRUE | 3.87 | 50 | 33 | 45 |
| TRUE | 3.86 | 52 | 36 | 39 |
| TRUE | 3.64 | 53 | 38 | 40 |
| TRUE | 3.59 | 53 | 34 | 40 |
The pivot_longer() and pivot_wider() functions rearrange data, decreasing or increasing the number of columns. The use of this will become more evident during visualization.
experiment %>%
filter(low.birthweight == TRUE) %>%
select(smoker, `miR-16`, `miR-21`, `miR-146a`, `miR-182`) %>%
pivot_longer(cols = c(`miR-16`, `miR-21`, `miR-146a`, `miR-182`),
names_to = "gene",
values_to = "expression") | smoker | gene | expression |
|---|---|---|
| TRUE | miR-16 | 60 |
| TRUE | miR-21 | 48 |
| TRUE | miR-146a | 89 |
| TRUE | miR-182 | 65 |
| TRUE | miR-16 | 70 |
| TRUE | miR-21 | 51 |
| TRUE | miR-146a | 84 |
| TRUE | miR-182 | 48 |
| TRUE | miR-16 | 61 |
| TRUE | miR-21 | 51 |
| TRUE | miR-146a | 97 |
| TRUE | miR-182 | 59 |
| TRUE | miR-16 | 47 |
| TRUE | miR-21 | 40 |
| TRUE | miR-146a | 87 |
| TRUE | miR-182 | 63 |
| FALSE | miR-16 | 64 |
| FALSE | miR-21 | 47 |
| FALSE | miR-146a | 104 |
| FALSE | miR-182 | 42 |
| TRUE | miR-16 | 61 |
| TRUE | miR-21 | 47 |
| TRUE | miR-146a | 77 |
| TRUE | miR-182 | 60 |
7.5 Exercise 4: converting between base R and Tidyverse
Reproduce the table 7.5 or table 7.6 using base R. Use Tidyverse functions to answer the question you addressed in exercise 3.