Immune profiling V(D)J with 10X
10x Genomics provides V(D)J references and T cell receptor (TCR) and B cell receptor (BCR) amplification kits for human and mouse. For other organisms, you will need to create your own primer set and reference sequence.
Library preparation
V(D)J library preparation occurs in five steps:
- GEM generation and barcoding Cells are partitioned in gel beads, then lysed. Full length cDNA from poly-adenylated mRNA is generated with an Illumina read 1 sequencing primer, a 16 nt 10x (cell) Barcode, a 10 nt unique molecular identifier (UMI), and a 13 nt template switching oligo (TSO) added to each molecule.
- GEM pooling and cleanup The cDNA from all the gel beads is pooled, and leftover reagents and primers are removed with a magnetic bead purification.
- cDNA amplification The cDNA is amplified to generate enough material to use in several libraries (e.g. V(D)J and GEX) from the same set of cells.
- Target enrichment TCR- or BCR-specific primers are used to enrich for V(D)J segments. P5 is added in this step.
- Library construction Fragmentation and size selection generate variable length fragments, and Illumina read 2 primer and P7 are added. Completed libraries have both P5 and P7, and are ready for sequencing.
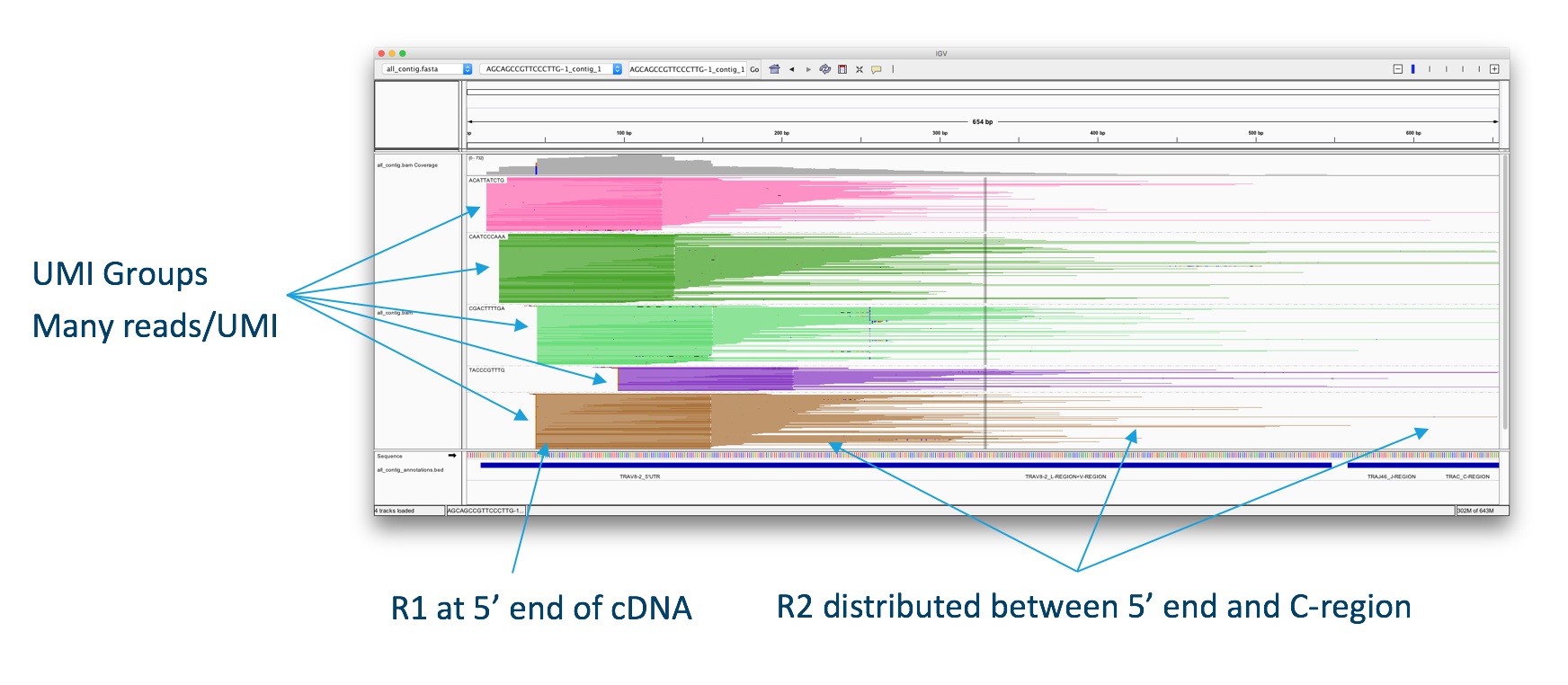
The V(D)J Algorithm
The figure above shows alignments of sequenced V(D)J read-pairs are aligned to an assembled chain, illustrating the structure of the read data. Thanks to the targeted enrichment followed by fragmentation, each transcript (UMI) may be represented by many reads with different insert lengths, resulting in different R2 start points. The diversity of R2 start points gives complete coverage of the targeted portion of each transcript, which is typically ~650bp.
The recommended sequencing depth for V(D)J libraries is ~5,000 reads per cell, 1/4 the recommendation for gene expression libraries.
Example data set
The data set we will be using in this workshop is from a 2021 study examining partial T-Cell immunity to COVID-19 induced by MMR and Tdap vaccination. In the data reduction section of this workshop, we will be using a single sample for the purposes of demonstration. When we reach the analysis portion of the workshop, this afternoon, we will pair the V(D)J data with the matched gene expression data generated in the experiment, as well.
The graphic below provides an overview of the experimental design:

Source: Mysore V, Cullere X, Settles ML, Ji X, Kattan MW, Desjardins M, Durbin-Johnson B, Gilboa T, Baden LR, Walt DR, Lichtman AH, Jehi L, Mayadas TN. Protective heterologous T cell immunity in COVID-19 induced by the trivalent MMR and Tdap vaccine antigens. Med (N Y). 2021 Sep 10;2(9):1050-1071.e7.
Data reduction
Log into tadpole and navigate to your directory on the /share/workshop space.
mkdir -p /share/workshop/vdj_workshop/$USER
cd /share/workshop/vdj_workshop/$USER
Request an interactive session from the scheduler so that we are not competing for resources on the head node.
srun -t 1-00:00:00 -c 4 -n 1 --mem 16000 --partition production --account workshop --reservation rnaworkshop --pty /bin/bash
Project set-up
Reads
We will be using a small fastq file that was created by subsetting 1,000,000 reads from the original data.
Create a symbolic link to the sequence data inside your project directory.
mkdir -p /share/workshop/vdj_workshop/$USER/00-RawData
cd /share/workshop/vdj_workshop/$USER/00-RawData
ln -s /share/workshop/vdj_workshop/Data/* .
Software
Before getting started, we need to make sure that we have the cellranger software in our path. This can be done one of three ways:
- Module load:
module load cellranger. This will only work on a cluster with modules for software management. - Add the location of a previously downloaded cellranger build to our path:
export PATH=/share/workshop/vdj_workshop/Software/cellranger-7.1.0/bin:$PATH. This will only work if there is a download of cellranger somewhere in the filesystem for you to point to. - Download cellranger in the current directory.
Add cellranger to the path.
export PATH=/share/workshop/vdj_workshop/Software/cellranger-7.1.0/bin:$PATH
Reference
There are prebuilt human and mouse reference packages for use with Cell Ranger V(D)J, which we will be using in this workshop. For other species, or to create a custom reference, we would run cellranger mkvdjref.
cellranger mkvdjref
The following code will generate a reference with cellranger mkvdjref. This takes a while, and is not used in this workshop. When using this code, please ensure that your FASTA and GTF files are appropriate versions, downloading up to date files as necessary.
cd /share/workshop/vdj_workshop/$USER/
cellranger mkvdjref \
--genome=reference \
--fasta=/share/genomes/ensembl/GRCh38/Homo_sapiens.GRCh38.dna_sm.primary_assembly.fa \
--genes=/share/genomes/ensembl/GRCh38/Homo_sapiens.GRCh38.89.gtf
This assumes that the following biotypes are present in the gtf files:
- TR_C_gene
- TR_D_gene
- TR_J_gene
- TR_V_gene
- IG_C_gene
- IG_D_gene
- IG_J_gene
- IG_V_gene
You can also generate V(D)J references for IMGT sequences. Additional instructions for building VDJ references can be found here.
Downloading prebuilt Cell Ranger reference
Since we are working with human data in this workshop, we will use a prebuilt reference that was downloaded from the 10x website using code available here.
Create a symbolic link to the downloaded prebuilt reference.
cd /share/workshop/vdj_workshop/$USER
ln -s /share/workshop/vdj_workshop/Software/refdata-cellranger-vdj-GRCh38-alts-ensembl-7.1.0 reference
Running cellranger vdj
For experiments with V(D)J libraries only, 10x recommends running cellranger vdj, while cellranger multi allows gene expression, V(D)J, and / or feature barcode libraries from the same experiment to be processed simultaneously. Detailed descriptions of cellranger vdj and cellranger multi can be found on the 10x website.
Input
The input to cellranger vdj is simple. The user is required to provide the path to a directory containing fastq files, the a sample identifier, and the path to a reference.
Launch cellranger vdj.
mkdir -p /share/workshop/vdj_workshop/$USER/01-Cellranger
cd /share/workshop/vdj_workshop/$USER/01-Cellranger
cellranger vdj \
--id=Pool1_VDJ \
--fastqs=/share/workshop/vdj_workshop/$USER/00-RawData \
--sample=Pool1_VDJ \
--reference=/share/workshop/vdj_workshop/$USER/reference
In this workshop, because we’re working on an interactive session with a single sample, we will run the command directly on the command line. A SLURM script to run an array of cellranger vdj jobs through a scheduler and a bash script to run the cellranger vdj command are provided in the Software directory for your reference.
Output
There are a lot of files created in the output folder, including:
- web_summary.html - similar to gene expression
- metrics_summary.csv - similar to gene expression
- annotation CSV/JSONs - filtered_contig_annotations.csv, clonotypes.csv
- FASTQ/FASTAs - filtered_contig.fasta/filtered_contig.fastq
- barcoded BAMs - consensus alignment mapping files
- cell_barcodes.json - barcodes which are identified as targeted cells.
Running cellranger multi
Cell Ranger Multi requires a little bit more set-up in the form of a config file, described here.
The Multi configuration file (not a true CSV) describes the locations of the references and libraries for the experiment. Use a command line text editor to create a file called “Pool1_multi.csv” containing the required information.
When you have the config file, you can launch cellranger multi:
cellranger multi --id=Pool1_VDJ --csv=Pool1_multi.csv
Prepare for Analysis
Finally, please download the Rmarkdown document that contains the code we’ll be using as the basis of our analysis this afternoon.
