Running UCD Bioinformatics Supernova Pipeline with Snakemake
HOW TO CALL THE SNAKEMAKE FILE (GENERAL):
snakemake -s snakemake.py **RULE_NAME**- Files needed:
snakemake.py,templates/cluster.json,templates/keith.json - Be sure to specify
running_locallyin thetemplate/keith.jsonfile.- If
running_locally== ‘True’:templates/cluster.jsonis not needed if running locally only.- don’t need to worry about the sbatch parameters in the
templates/keith.jsonfile - Follow rules under RUNNING LOCALLY below.
- Else if
running_locally== ‘False’:- be sure to set all sbatch parameters in the
templates/keith.jsonfile - Follow rules under RUNNING ON CLUSTER below.
- be sure to set all sbatch parameters in the
- If
RUNNING ON CLUSTER
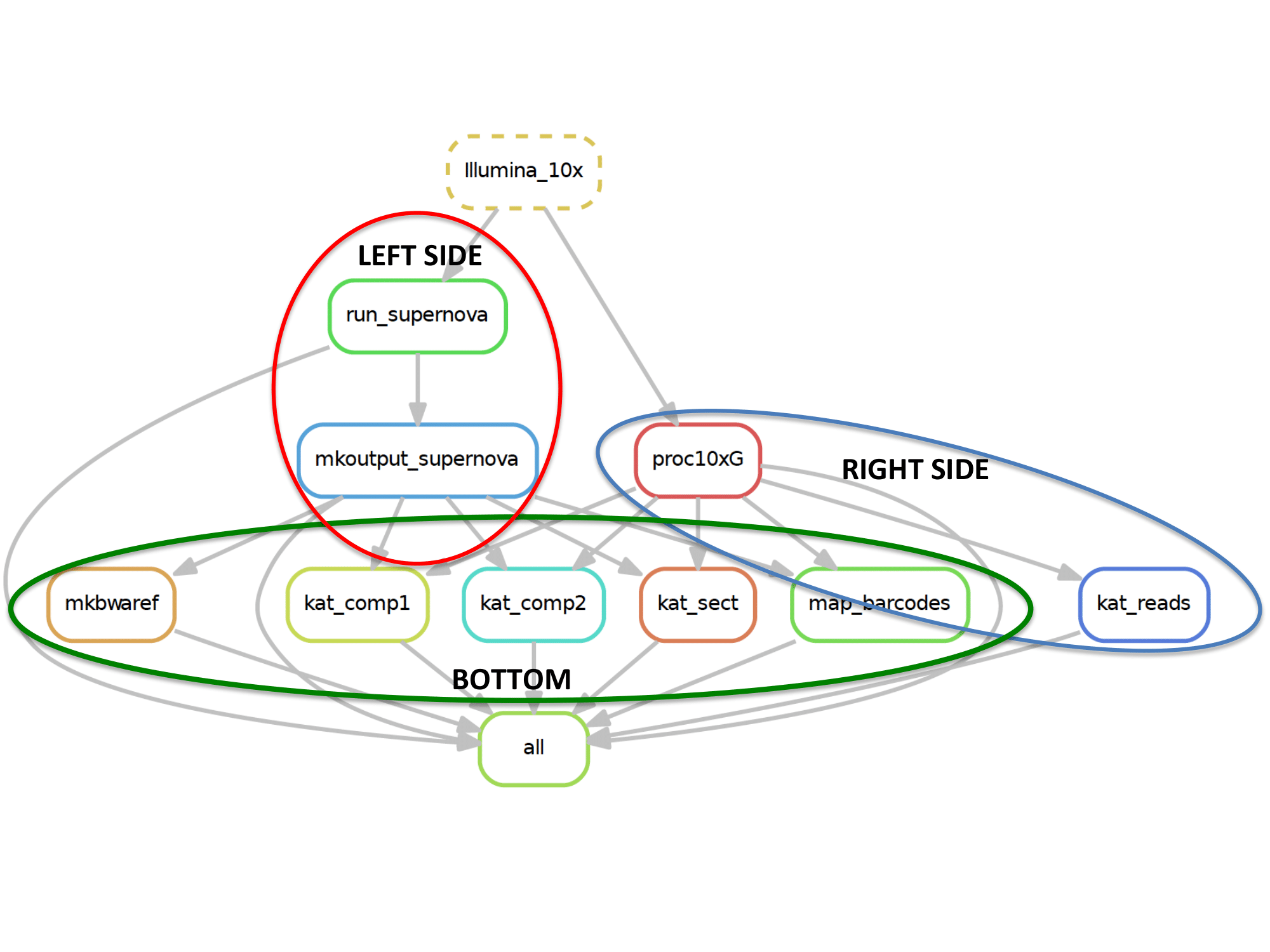
- Left Side
- running_locally = “False”, sbatch included in snakemake call and run supernova runs on that job and call sbatch for mkoutput (3x)
snakemake -s snakemake.py --cluster-config templates/cluster.json --cluster "sbatch -J {cluster.ls.job-name} -N {cluster.ls.n} -p {cluster.ls.partition} -t {cluster.ls.time} -m {cluster.ls.mem} --output {cluster.ls.output} --error {cluster.ls.error} --mail-type {cluster.ls.mail-type} --mail-user {cluster.ls.mail-user} --exclusive" left_side
- Right Side
- running_locally = “False”, sbatch included in snakemake call and proc10xg runs on that job and call sbatch for kat reads (3x)
snakemake -s snakemake.py --cluster-config templates/cluster.json --cluster "sbatch -J {cluster.rs.job-name} -N {cluster.rs.n} -p {cluster.rs.partition} -t {cluster.rs.time} -n {cluster.rs.ntasks} -m {cluster.rs.mem} --output {cluster.rs.output} --error {cluster.rs.error} --mail-type {cluster.rs.mail-type} --mail-user {cluster.rs.mail-user}" right_side
- Bottom
- running_locally = “False”, no sbatch included in call or included (doesnt matter) all portions submitted as an sbatch
snakemake -s snakemake.py bottom
- Running 1 rule of interest?
- running_locally = “False”,
- All rules except
run_supernovaandproc10xGsnakemake -s snakemake.py **RULE_NAME**
run_supernova- Run number 1, except replace
left_sidewithrun_supernovaat the end of the command.
- Run number 1, except replace
proc10xG- Run number 2, except replace
right_sidewithproc10xGat the end of the command.
- Run number 2, except replace
RUNNING LOCALLY (TODO)
- All
- running_locally = “True”
snakemake -s snakemake.py all
- Left Side
- running_locally = “True”
snakemake -s snakemake.py all
- Right Side
- running_locally = “True”
snakemake -s snakemake.py all
- Bottom
- running_locally = “True”
snakemake -s snakemake.py all
- Running 1 rule of interest? (IDK ABOUT THIS QUITE YET)
- running_locally = “True”, no sbatch included in call
snakemake -s snakemake.py **RULE_NAME*****
