Last Updated: March 23 2021, 5pm
Part 1: Loading data from CellRanger into R
Our first Markdown document concentrates on getting data into R and setting up our initial object.
Single Cell Analysis with Seurat and some custom code!
Seurat (now Version 4) is a popular R package that is designed for QC, analysis, and exploration of single cell data. Seurat aims to enable users to identify and interpret sources of heterogeneity from single cell transcriptomic measurements, and to integrate diverse types of single cell data. Further, the authors provide several tutorials, on their website.
The intro2singlecell_March2021.zip file contains the single cell matrix files and HDF5 files for four Covid positive PBMc samples. These are PBMC human cells ran on the 10X genomics platform (5’ gene expression kit V2 with b-cell VDJ) for single cell RNA sequencing, sequenced with UC Davis in Nov and Dec of 2020 on a NovaSeq 6000. The experiment for the workshop contains 4 samples, each merged from 2 original samples and “normal” PBMC data from 10X Genomics.
The four samples are,
- PBMC2 - Performed in Nov
- PBMC3 - Performed in Nov
- T021 - Performed in Dec
- T022 - Performed in Dec
Also in the directory is the 10X “normal” PBMC dataset
We start each markdown document with loading needed libraries for R
# must have Seurat
library(Seurat)
library(kableExtra)
library(ggplot2)
Setup the experiment folder and data info
experiment_name = "Covid Example"
dataset_loc <- "./intro2singlecell_March2021"
ids <- c("PBMC2", "PBMC3", "T021PBMC", "T022PBMC")
Read in the cellranger sample metrics csv files
d10x.metrics <- lapply(ids, function(i){
# remove _Counts is if names don't include them
metrics <- read.csv(file.path(dataset_loc,paste0(i,"_Counts/outs"),"metrics_summary.csv"), colClasses = "character")
})
experiment.metrics <- do.call("rbind", d10x.metrics)
rownames(experiment.metrics) <- ids
sequencing_metrics <- data.frame(t(experiment.metrics[,c(4:17,1,18,2,3,19,20)]))
row.names(sequencing_metrics) <- gsub("\\."," ", rownames(sequencing_metrics))
And lets generate a pretty table
sequencing_metrics %>%
kable(caption = 'Cell Ranger Results') %>%
pack_rows("Sequencing Characteristics", 1, 7, label_row_css = "background-color: #666; color: #fff;") %>%
pack_rows("Mapping Characteristics", 8, 14, label_row_css = "background-color: #666; color: #fff;") %>%
pack_rows("Cell Characteristics", 15, 20, label_row_css = "background-color: #666; color: #fff;") %>%
kable_styling("striped")
| PBMC2 | PBMC3 | T021PBMC | T022PBMC | |
|---|---|---|---|---|
| Sequencing Characteristics | ||||
| Number of Reads | 313,087,958 | 339,008,152 | 364,881,065 | 358,269,446 |
| Valid Barcodes | 85.1% | 82.0% | 91.5% | 91.0% |
| Sequencing Saturation | 78.8% | 76.5% | 85.2% | 87.3% |
| Q30 Bases in Barcode | 97.2% | 97.1% | 94.3% | 94.4% |
| Q30 Bases in RNA Read | 94.5% | 94.5% | 87.3% | 87.4% |
| Q30 Bases in RNA Read 2 | 91.1% | 91.1% | 89.3% | 88.7% |
| Q30 Bases in UMI | 96.9% | 96.9% | 93.9% | 94.0% |
| Mapping Characteristics | ||||
| Reads Mapped to Genome | 91.2% | 91.3% | 90.9% | 90.0% |
| Reads Mapped Confidently to Genome | 70.0% | 69.4% | 83.5% | 82.9% |
| Reads Mapped Confidently to Intergenic Regions | 5.8% | 5.5% | 4.1% | 4.4% |
| Reads Mapped Confidently to Intronic Regions | 8.9% | 7.3% | 6.6% | 6.9% |
| Reads Mapped Confidently to Exonic Regions | 55.3% | 56.7% | 72.9% | 71.6% |
| Reads Mapped Confidently to Transcriptome | 48.6% | 48.2% | 67.3% | 65.6% |
| Reads Mapped Antisense to Gene | 4.3% | 6.5% | 3.0% | 3.4% |
| Cell Characteristics | ||||
| Estimated Number of Cells | 6,914 | 7,483 | 7,891 | 5,120 |
| Fraction Reads in Cells | 53.2% | 89.5% | 92.0% | 92.6% |
| Mean Reads per Cell | 45,283 | 45,304 | 46,240 | 69,975 |
| Median Genes per Cell | 840 | 1,385 | 1,506 | 1,739 |
| Total Genes Detected | 20,425 | 20,681 | 21,012 | 20,774 |
| Median UMI Counts per Cell | 1,506 | 3,384 | 3,912 | 4,736 |
Load the Cell Ranger Matrix Data and create the base Seurat object.
Cell Ranger provides a function cellranger aggr that will combine multiple samples into a single matrix file. However, when processing data in R this is unnecessary and we can quickly aggregate them in R.
Seurat provides a function Read10X and Read10X_h5 to read in 10X data folder. First we read in data from each individual sample folder.
Later, we initialize the Seurat object (CreateSeuratObject) with the raw (non-normalized data). Keep all cells with at least 200 detected genes. Also extracting sample names, calculating and adding in the metadata mitochondrial percentage of each cell. Adding in the metadata batchid and cell cycle. Finally, saving the raw Seurat object.
Load the Cell Ranger Matrix Data (hdf5 file) and create the base Seurat object.
d10x.data <- lapply(ids, function(i){
d10x <- Read10X_h5(file.path(dataset_loc,paste0(i,"_Counts/outs"),"raw_feature_bc_matrix.h5"))
colnames(d10x) <- paste(sapply(strsplit(colnames(d10x),split="-"),'[[',1L),i,sep="-")
d10x
})
names(d10x.data) <- ids
str(d10x.data)
If you don’t have the needed hdf5 libraries you can read in the matrix files like such
d10x.data <- sapply(ids, function(i){
d10x <- Read10X(file.path(dataset_loc,paste0(i,"_Counts/outs"),"raw_feature_bc_matrix"))
colnames(d10x) <- paste(sapply(strsplit(colnames(d10x),split="-"),'[[',1L),i,sep="-")
d10x
})
names(d10x.data) <- ids
Lets recreate the pretty cellranger html plot
cr_filtered_cells <- as.numeric(gsub(",","",as.character(unlist(sequencing_metrics["Estimated Number of Cells",]))))
plot_cellranger_cells <- function(ind){
xbreaks = c(1,1e1,1e2,1e3,1e4,1e5,1e6)
xlabels = c("1","10","100","1000","10k","100K","1M")
ybreaks = c(1,2,5,10,20,50,100,200,500,1000,2000,5000,10000,20000,50000,100000,200000,500000,1000000)
ylabels = c("1","2","5","10","2","5","100","2","5","1000","2","5","10k","2","5","100K","2","5","1M")
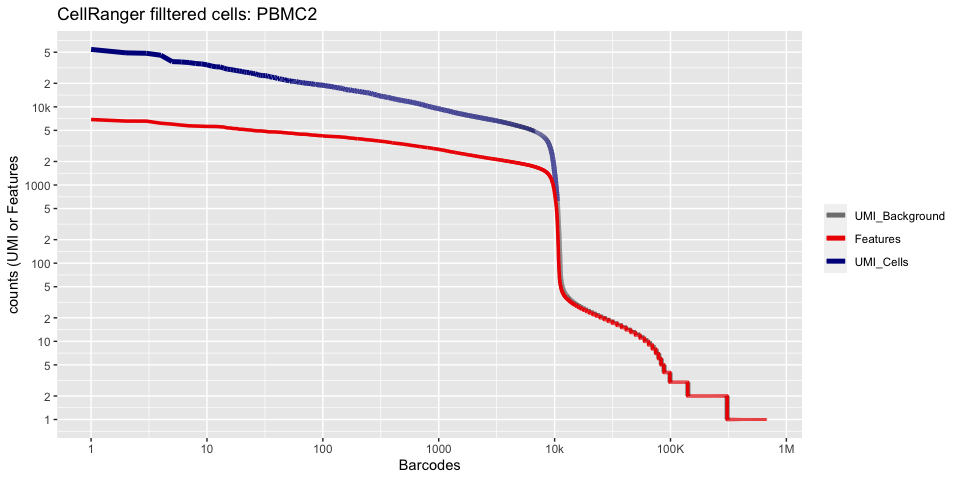
pl1 <- data.frame(index=seq.int(1,ncol(d10x.data[[ind]])), nCount_RNA = sort(Matrix:::colSums(d10x.data[[ind]])+1,decreasing=T), nFeature_RNA = sort(Matrix:::colSums(d10x.data[[ind]]>0)+1,decreasing=T)) %>% ggplot() +
scale_color_manual(values=c("grey50","red2","blue4"), labels=c("UMI_Background", "Features", "UMI_Cells"), name=NULL) +
ggtitle(paste("CellRanger filltered cells:",ids[ind],sep=" ")) + xlab("Barcodes") + ylab("counts (UMI or Features") +
scale_x_continuous(trans = 'log2', breaks=xbreaks, labels = xlabels) +
scale_y_continuous(trans = 'log2', breaks=ybreaks, labels = ylabels) +
geom_line(aes(x=index, y=nCount_RNA, color=index<=cr_filtered_cells[ind] , group=1), size=1.75) +
geom_line(aes(x=index, y=nFeature_RNA, color="Features", group=1), size=1.25)
return(pl1)
}
plot_cellranger_cells(1)
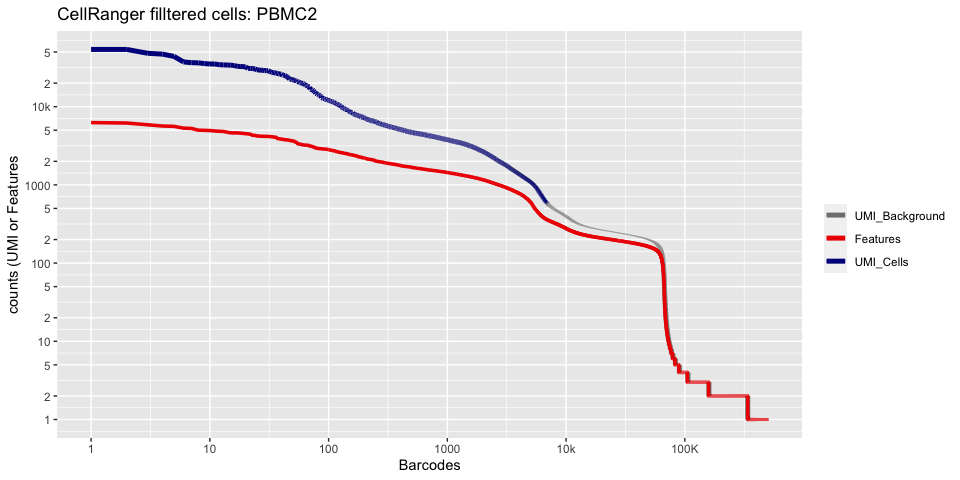
plot_cellranger_cells(2)
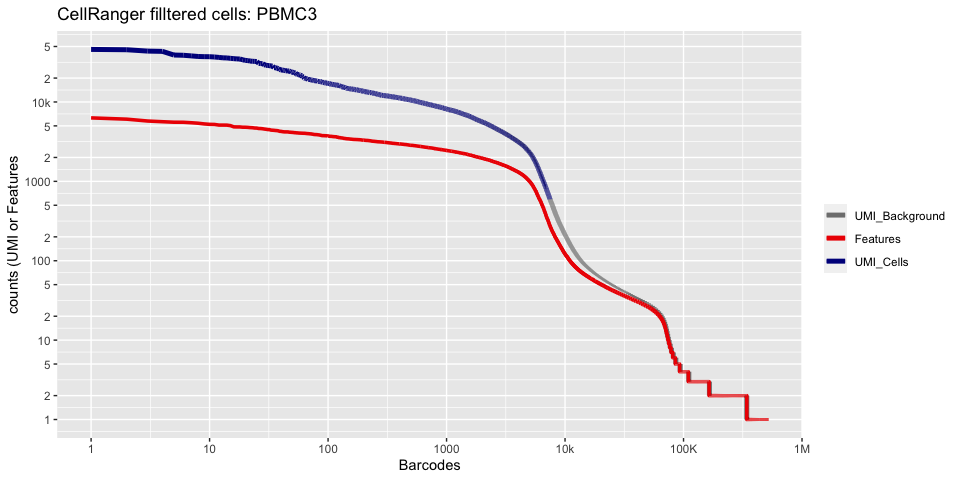
plot_cellranger_cells(3)
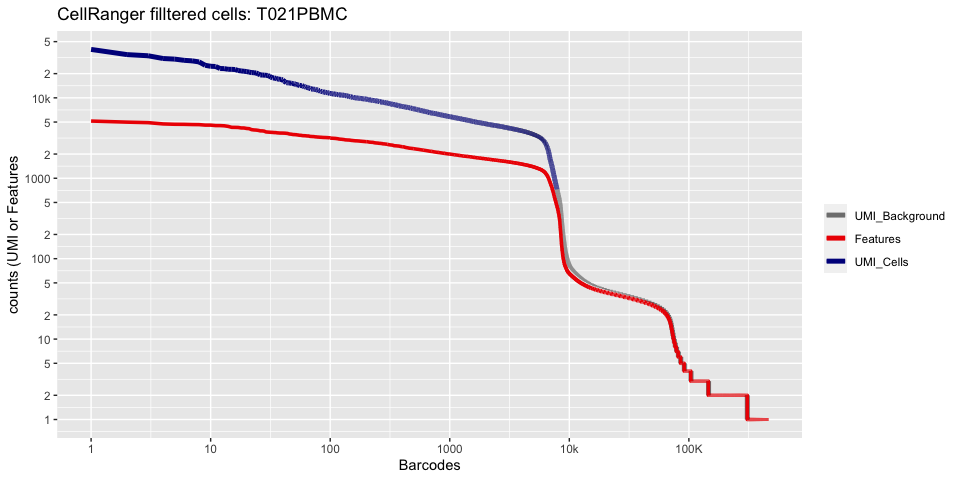
plot_cellranger_cells(4)
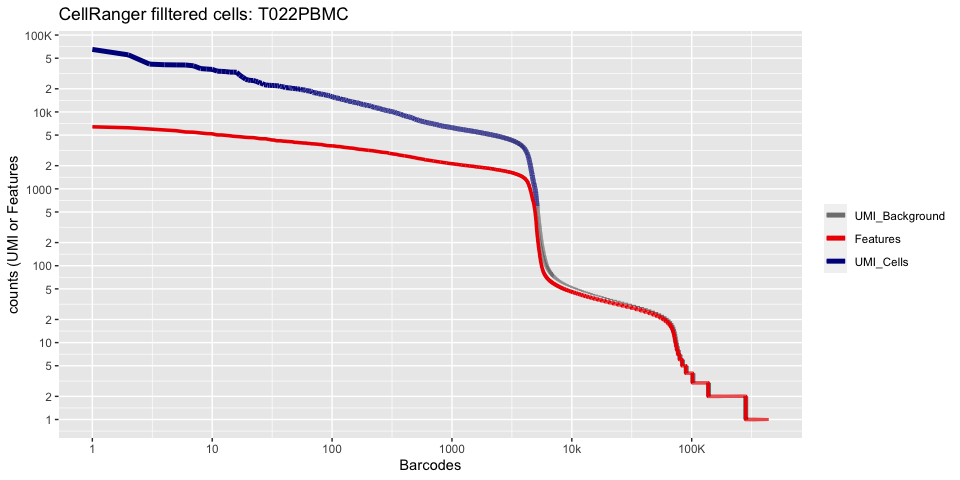
Create the Seurat object
filter criteria: remove genes that do not occur in a minimum of 0 cells and remove cells that don’t have a minimum of 200 features
experiment.data <- do.call("cbind", d10x.data)
experiment.aggregate <- CreateSeuratObject(
experiment.data,
project = experiment_name,
min.cells = 0,
min.features = 300,
names.field = 2,
names.delim = "\\-")
experiment.aggregate
str(experiment.aggregate)
Load the Cell Ranger Matrix Data with multiple tables (hdf5 file only) and create the base Seurat object.
normals <- c("10x_NormalPBMC_multi")
d10x.metrics <- lapply(normals, function(i){
# remove _Counts is my names don't include them
metrics <- read.csv(file.path(dataset_loc,paste0(i,"/outs"),"metrics_summary.csv"), colClasses = "character")
metrics <- metrics[metrics$Library.Type == "Gene Expression",]
metrics.new <- metrics$Metric.Value
names(metrics.new) <- metrics$Metric.Name
metrics.new
})
normal.metrics <- do.call("rbind", d10x.metrics)
rownames(normal.metrics) <- normals
d10x.normal <- lapply(normals, function(i){
d10x <- Read10X_h5(file.path(dataset_loc,paste0(i,"/outs"),"raw_feature_bc_matrix.h5"))
colnames(d10x$`Gene Expression`) <- paste(sapply(strsplit(colnames(d10x$`Gene Expression`),split="-"),'[[',1L),i,sep="-")
colnames(d10x$`Antibody Capture`) <- paste(sapply(strsplit(colnames(d10x$`Antibody Capture`),split="-"),'[[',1L),i,sep="-")
d10x
})
names(d10x.normal) <- normals
str(d10x.normal)
How does d10x.data differ from d10x.normal?
cr_filtered_cells <- as.numeric(gsub(",","",as.character(unlist(normal.metrics[, "Estimated number of cells" ]))))
plot_cellranger_cells <- function(ind){
xbreaks = c(1,1e1,1e2,1e3,1e4,1e5,1e6)
xlabels = c("1","10","100","1000","10k","100K","1M")
ybreaks = c(1,2,5,10,20,50,100,200,500,1000,2000,5000,10000,20000,50000,100000,200000,500000,1000000)
ylabels = c("1","2","5","10","2","5","100","2","5","1000","2","5","10k","2","5","100K","2","5","1M")
pl1 <- data.frame(index=seq.int(1,ncol(d10x.normal[[ind]][["Gene Expression"]])), nCount_RNA = sort(Matrix:::colSums(d10x.normal[[ind]][["Gene Expression"]])+1,decreasing=T), nFeature_RNA = sort(Matrix:::colSums(d10x.normal[[ind]][["Gene Expression"]]>0)+1,decreasing=T)) %>% ggplot() +
scale_color_manual(values=c("grey50","red2","blue4"), labels=c("UMI_Background", "Features", "UMI_Cells"), name=NULL) +
ggtitle(paste("CellRanger filltered cells:",ids[ind],sep=" ")) + xlab("Barcodes") + ylab("counts (UMI or Features") +
scale_x_continuous(trans = 'log2', breaks=xbreaks, labels = xlabels) +
scale_y_continuous(trans = 'log2', breaks=ybreaks, labels = ylabels) +
geom_line(aes(x=index, y=nCount_RNA, color=index<=cr_filtered_cells[ind] , group=1), size=1.75) +
geom_line(aes(x=index, y=nFeature_RNA, color="Features", group=1), size=1.25)
return(pl1)
}
plot_cellranger_cells(1)
Create the Seurat object
filter criteria: remove genes that do not occur in a minimum of 0 cells and remove cells that don’t have a minimum of 200 features
experiment.normal <- do.call("cbind", lapply(d10x.normal,"[[", "Gene Expression"))
experiment.aggregate.normal <- CreateSeuratObject(
experiment.normal,
project = "Normals",
min.cells = 0,
min.features = 300,
names.field = 2,
names.delim = "\\-")
experiment.aggregate
str(experiment.aggregate)
Lets create a metadata variable call batch, based on the sequencing run
Here we build a new metadata variable ‘batchid’ which can be used to specify treatment groups.
samplename = experiment.aggregate$orig.ident
batchid = rep("Batch1",length(samplename))
batchid[samplename %in% c("T021PBMC","T022PBMC")] = "Batch2"
names(batchid) = colnames(experiment.aggregate)
experiment.aggregate <- AddMetaData(
object = experiment.aggregate,
metadata = batchid,
col.name = "batchid")
table(experiment.aggregate$batchid)
The percentage of reads that map to the mitochondrial genome
- Low-quality / dying cells often exhibit extensive mitochondrial contamination.
- We calculate mitochondrial QC metrics with the PercentageFeatureSet function, which calculates the percentage of counts originating from a set of features.
- We use the set of all genes, in mouse these genes can be identified as those that begin with ‘mt’, in human data they begin with MT.
experiment.aggregate$percent.mito <- PercentageFeatureSet(experiment.aggregate, pattern = "^MT-")
summary(experiment.aggregate$percent.mito)
Lets spend a little time getting to know the Seurat object.
The Seurat object is the center of each single cell analysis. It stores all information associated with the dataset, including data, annotations, analyses, etc. The R function slotNames can be used to view the slot names within an object.
slotNames(experiment.aggregate)
head(experiment.aggregate[[]])
Question(s)
- What slots are empty, what slots have data?
- What columns are available in meta.data?
- Look up the help documentation for subset?
Finally, save the original object and view the object.
Original dataset in Seurat class, with no filtering
save(experiment.aggregate,file="original_seurat_object.RData")
save(experiment.aggregate.normal,file="normal_seurat_object.RData")
Get the next Rmd file
download.file("https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2021-August-Single-Cell-RNA-Seq-Analysis/master/data_analysis/scRNA_Workshop-PART2.Rmd", "scRNA_Workshop-PART2.Rmd")
Session Information
sessionInfo()
