Spatial Transcriptomics Part 4: Simply Niche analysis
Set up the workspace
library(Seurat) # Spatial transcriptomics analysis
library(kableExtra) # format tables
library(ggplot2) # create graphics
library(viridis) # accessible color palettes
library(ComplexHeatmap) # visualization heatmaps
We are going to use the cortex region that was subsetted previously for this exercise.
cortex <- readRDS("cortex.rds")
Find niches
Up until now, we have done analyses that are very similar to regular single cell RNASeq data analysis, without utilizing the spatial information of the cell except for in visualizations. One of the popular analyses for spatial transcriptomics is to identify niches. A nich are regions of tissue, each of which is defined by a different composition of spatially adjacent cell types. In Seurat, a local neighborhood for each cell is constructed by including its neighbors.k spatially closest neighbors, and count the occurences of each cell type present in this neighborhood. Then a k-means clustering is done to group cells that have similar neighborhoods together, which is defined as spatial niches.
We will use BuildNicheAssay function to perform this analysis.
cortex <- BuildNicheAssay(cortex, fov = "fov.TgCRND8", group.by = "predicted.celltype", niches.k = 5, neighbors.k = 30)
Let’s visualize the niche result together with the cell types.
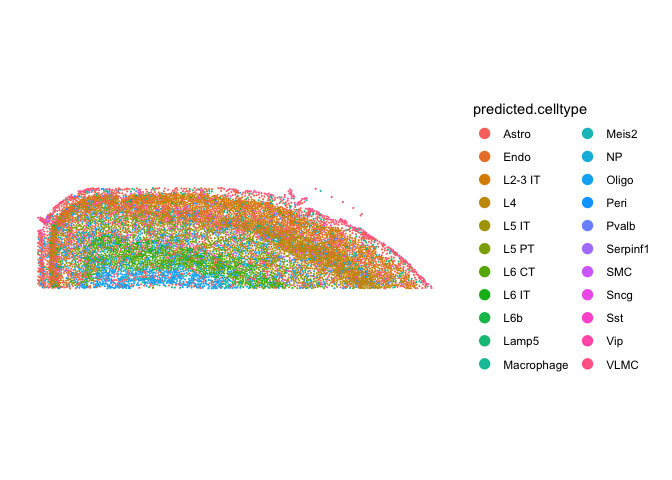
ImageDimPlot(cortex, fov = "fov.TgCRND8", group.by = "predicted.celltype", size = 0.75, dark.background = F)
ImageDimPlot(cortex, fov = "fov.TgCRND8", group.by = "niches", size = 0.75, dark.background = F)
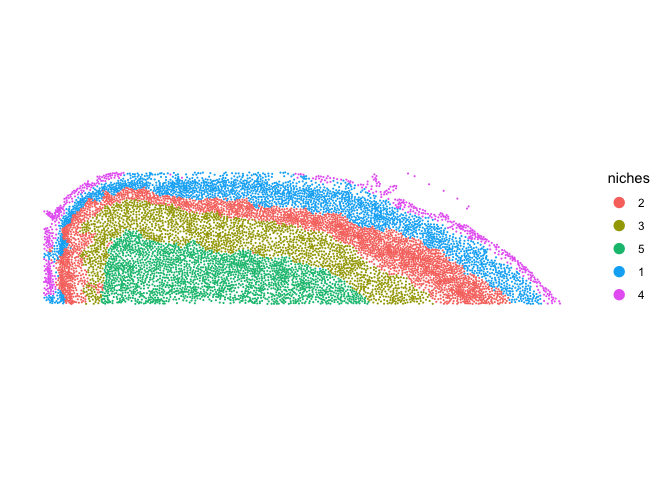
Next, we can tally the number of each cell type within a niche.
mat <- table(cortex$predicted.celltype, cortex$niches)
mat
##
## 1 2 3 4 5
## Astro 647 629 331 205 286
## Endo 138 154 117 46 76
## L2-3 IT 1033 84 3 0 3
## L4 130 1369 47 0 7
## L5 IT 3 78 349 1 30
## L5 PT 17 39 403 0 6
## L6 CT 3 6 27 0 718
## L6 IT 24 12 35 3 159
## L6b 0 1 6 0 63
## Lamp5 91 13 10 21 16
## Macrophage 102 115 90 25 92
## Meis2 0 0 0 0 1
## NP 0 1 85 0 6
## Oligo 27 172 174 19 677
## Peri 21 21 20 3 18
## Pvalb 38 81 92 0 31
## Serpinf1 8 2 1 0 1
## SMC 4 3 0 15 0
## Sncg 11 1 4 0 1
## Sst 34 33 80 0 14
## Vip 13 20 2 1 2
## VLMC 6 4 1 265 3
Let’s visualize this relationship
cell_fun = function(j, i, x, y, width, height, fill) {
grid::grid.rect(x = x, y = y, width = width *0.99,
height = height *0.99,
gp = grid::gpar(col = "grey",
fill = fill, lty = 1, lwd = 0.5))
}
col_fun=circlize::colorRamp2(c(-2, 0, 2), c("blue", "white", "red"))
Heatmap(scale(mat), show_row_dend = F, show_column_dend = F, rect_gp = grid::gpar(type = "none"), cell_fun = cell_fun, col = col_fun, column_names_rot = 90)
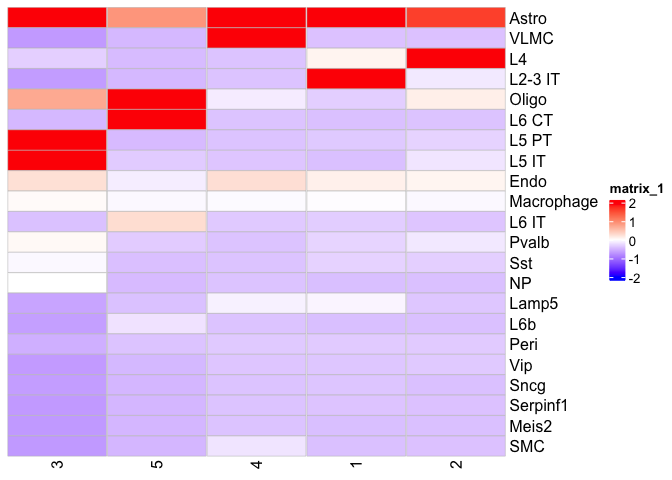
Please play around with the number of niches that BuildNicheAssay should find and see how the cell type distribution changes.
library(SpatialExperiment)
library(SummarizedExperiment)
library(scuttle)
library(scater)
library(cowplot)
cortex <- readRDS("cortex.rds")
exp <- LayerData(cortex, layer = "counts")
coords <- GetTissueCoordinates(cortex, which = "centroids")
rownames(coords) <- coords$cell
coords <- as.matrix(coords[,1:2])
se <- SpatialExperiment(assay = list(counts = exp), spatialCoords = coords)
se <- computeLibraryFactors(se)
assay(se, "data") <- normalizeCounts(se)
k_geom <- c(15, 30)
se <- computeBanksy(se, compute_agf = T, k_geom = k_geom, assay_name = "data")
lambda <- c(0.2, 0.8)
set.seed(1000)
se <- runBanksyPCA(se, use_agf = T, lambda = lambda)
se <- runBanksyUMAP(se, use_agf = T, lambda = lambda)
se <- clusterBanksy(se, use_agf = T, lambda = lambda, resolution = c(0.3, 0.5, 0.8, 1))
cnames <- colnames(colData(se))
cnames <- cnames[grep("^clust", cnames)]
colData(se) <- cbind(colData(se), spatialCoords(se))
plot_lam02res08 <- plotColData(se,
x = 'x', y = 'y',
point_size = 0.2, colour_by = cnames[1]
)
plot_lam02res1 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[2]
)
plot_lam08res08 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[3]
)
plot_lam08res1 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[4]
)
plot_lam02res03 <- plotColData(se,
x = 'x', y = 'y',
point_size = 0.2, colour_by = cnames[5]
)
plot_lam02res05 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[6]
)
plot_lam08res03 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[7]
)
plot_lam08res05 <- plotColData(se,
x = "x", y = "y",
point_size = 0.2, colour_by = cnames[8]
)
pdf("banksy.pdf", height = 30, width = 20)
plot_grid(plotlist = list(plot_lam02res08 + coord_equal(), plot_lam02res1 + coord_equal(),
plot_lam08res08 + coord_equal(), plot_lam08res1 + coord_equal(),
plot_lam02res03 + coord_equal(), plot_lam02res05 + coord_equal(),
plot_lam08res03 + coord_equal(), plot_lam08res05 + coord_equal()),
ncol = 1)
dev.off()
Prepare for the next section
Download Rmd
```{r download_Rmd, eval=FALSE} download.file(“https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2025-March-Spatial-Transcriptomics/main/data_analysis/05-NicheDE.Rmd”, “05-NicheDE.Rmd”)
#### Session information
```{r sessioinfo}
sessionInfo()
Prepare for the next section
Download Rmd
download.file("https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2025-March-Spatial-Transcriptomics/main/data_analysis/05-NicheDE.Rmd", "05-NicheDE.Rmd")
Session information
sessionInfo()
## R version 4.4.3 (2025-02-28)
## Platform: aarch64-apple-darwin20
## Running under: macOS Ventura 13.7.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/Los_Angeles
## tzcode source: internal
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] ComplexHeatmap_2.20.0 viridis_0.6.5 viridisLite_0.4.2
## [4] ggplot2_3.5.1 kableExtra_1.4.0 Seurat_5.2.1
## [7] SeuratObject_5.0.2 sp_2.1-4
##
## loaded via a namespace (and not attached):
## [1] RColorBrewer_1.1-3 shape_1.4.6.1 rstudioapi_0.16.0
## [4] jsonlite_1.8.8 magrittr_2.0.3 magick_2.8.5
## [7] spatstat.utils_3.1-2 farver_2.1.2 rmarkdown_2.27
## [10] GlobalOptions_0.1.2 vctrs_0.6.5 ROCR_1.0-11
## [13] Cairo_1.6-2 spatstat.explore_3.2-7 htmltools_0.5.8.1
## [16] sass_0.4.9 sctransform_0.4.1 parallelly_1.37.1
## [19] KernSmooth_2.23-26 bslib_0.7.0 htmlwidgets_1.6.4
## [22] ica_1.0-3 plyr_1.8.9 plotly_4.10.4
## [25] zoo_1.8-12 cachem_1.1.0 igraph_2.0.3
## [28] mime_0.12 lifecycle_1.0.4 iterators_1.0.14
## [31] pkgconfig_2.0.3 Matrix_1.7-2 R6_2.5.1
## [34] fastmap_1.2.0 clue_0.3-65 fitdistrplus_1.1-11
## [37] future_1.33.2 shiny_1.8.1.1 digest_0.6.35
## [40] colorspace_2.1-0 S4Vectors_0.44.0 patchwork_1.2.0
## [43] tensor_1.5 RSpectra_0.16-1 irlba_2.3.5.1
## [46] labeling_0.4.3 progressr_0.14.0 fansi_1.0.6
## [49] spatstat.sparse_3.0-3 httr_1.4.7 polyclip_1.10-6
## [52] abind_1.4-5 compiler_4.4.3 withr_3.0.0
## [55] doParallel_1.0.17 fastDummies_1.7.3 highr_0.11
## [58] MASS_7.3-64 rjson_0.2.21 tools_4.4.3
## [61] lmtest_0.9-40 httpuv_1.6.15 future.apply_1.11.2
## [64] goftest_1.2-3 glue_1.7.0 nlme_3.1-167
## [67] promises_1.3.0 Rtsne_0.17 cluster_2.1.8
## [70] reshape2_1.4.4 generics_0.1.3 gtable_0.3.5
## [73] spatstat.data_3.0-4 tidyr_1.3.1 data.table_1.15.4
## [76] xml2_1.3.6 utf8_1.2.4 BiocGenerics_0.50.0
## [79] spatstat.geom_3.2-9 RcppAnnoy_0.0.22 ggrepel_0.9.5
## [82] RANN_2.6.1 foreach_1.5.2 pillar_1.9.0
## [85] stringr_1.5.1 spam_2.10-0 RcppHNSW_0.6.0
## [88] later_1.3.2 circlize_0.4.16 splines_4.4.3
## [91] dplyr_1.1.4 lattice_0.22-6 survival_3.8-3
## [94] deldir_2.0-4 tidyselect_1.2.1 miniUI_0.1.1.1
## [97] pbapply_1.7-2 knitr_1.47 gridExtra_2.3
## [100] IRanges_2.38.0 svglite_2.1.3 scattermore_1.2
## [103] stats4_4.4.3 xfun_0.44 matrixStats_1.3.0
## [106] stringi_1.8.4 lazyeval_0.2.2 yaml_2.3.8
## [109] evaluate_0.23 codetools_0.2-20 tibble_3.2.1
## [112] cli_3.6.2 uwot_0.2.2 xtable_1.8-4
## [115] reticulate_1.39.0 systemfonts_1.1.0 munsell_0.5.1
## [118] jquerylib_0.1.4 Rcpp_1.0.12 globals_0.16.3
## [121] spatstat.random_3.2-3 png_0.1-8 parallel_4.4.3
## [124] dotCall64_1.1-1 listenv_0.9.1 scales_1.3.0
## [127] ggridges_0.5.6 purrr_1.0.2 crayon_1.5.2
## [130] GetoptLong_1.0.5 rlang_1.1.3 cowplot_1.1.3
