Retreiving Annotation via biomart
Annotation from BioMart with Ensembl names is most flexible way to retrieve tabular annotation for an organism.
- The Biomart start page should look like …
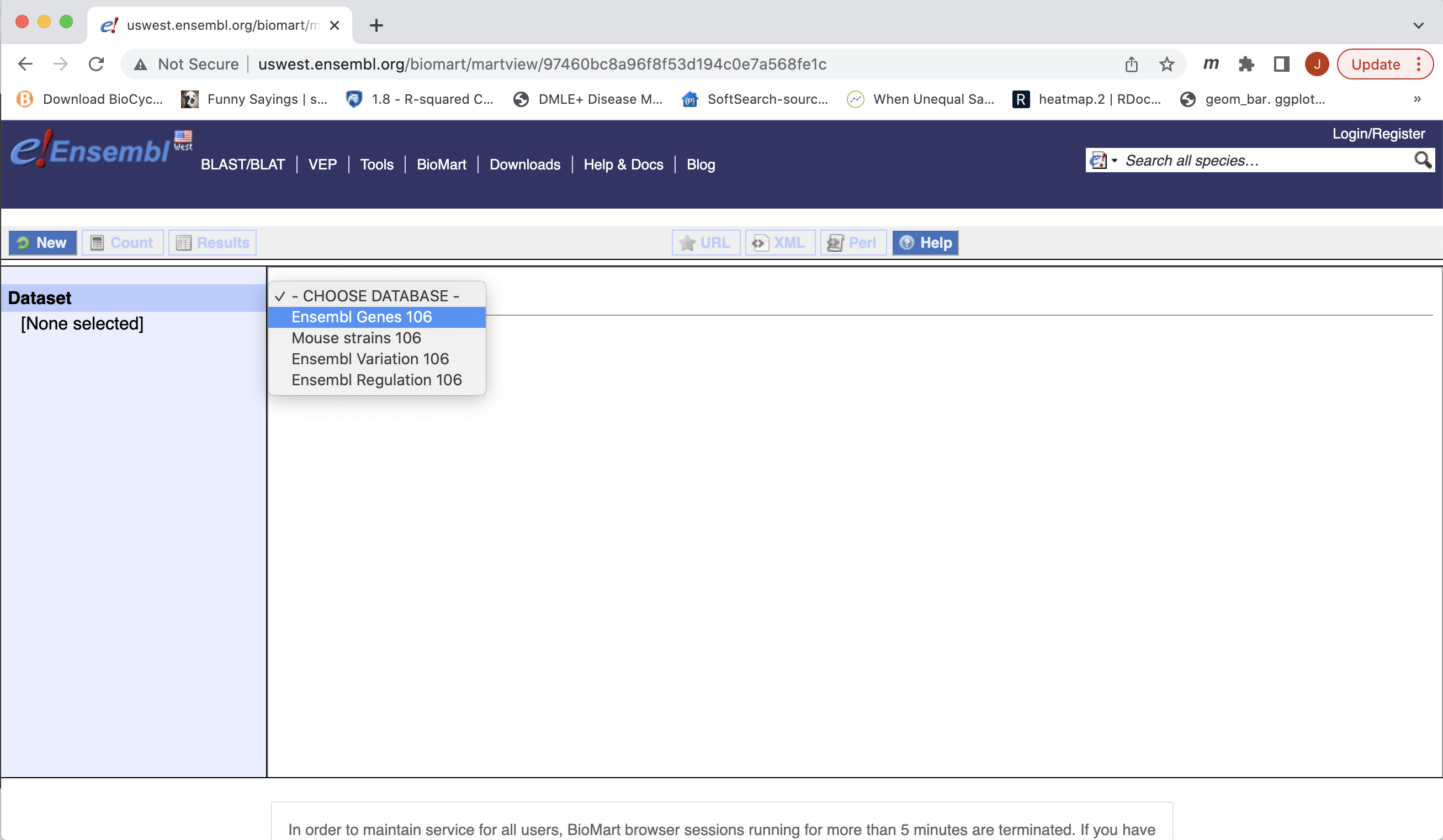
- First select the dataset, for gene expression experiment select Ensembl Genes 106 (version 106). The current version as of this workshop.
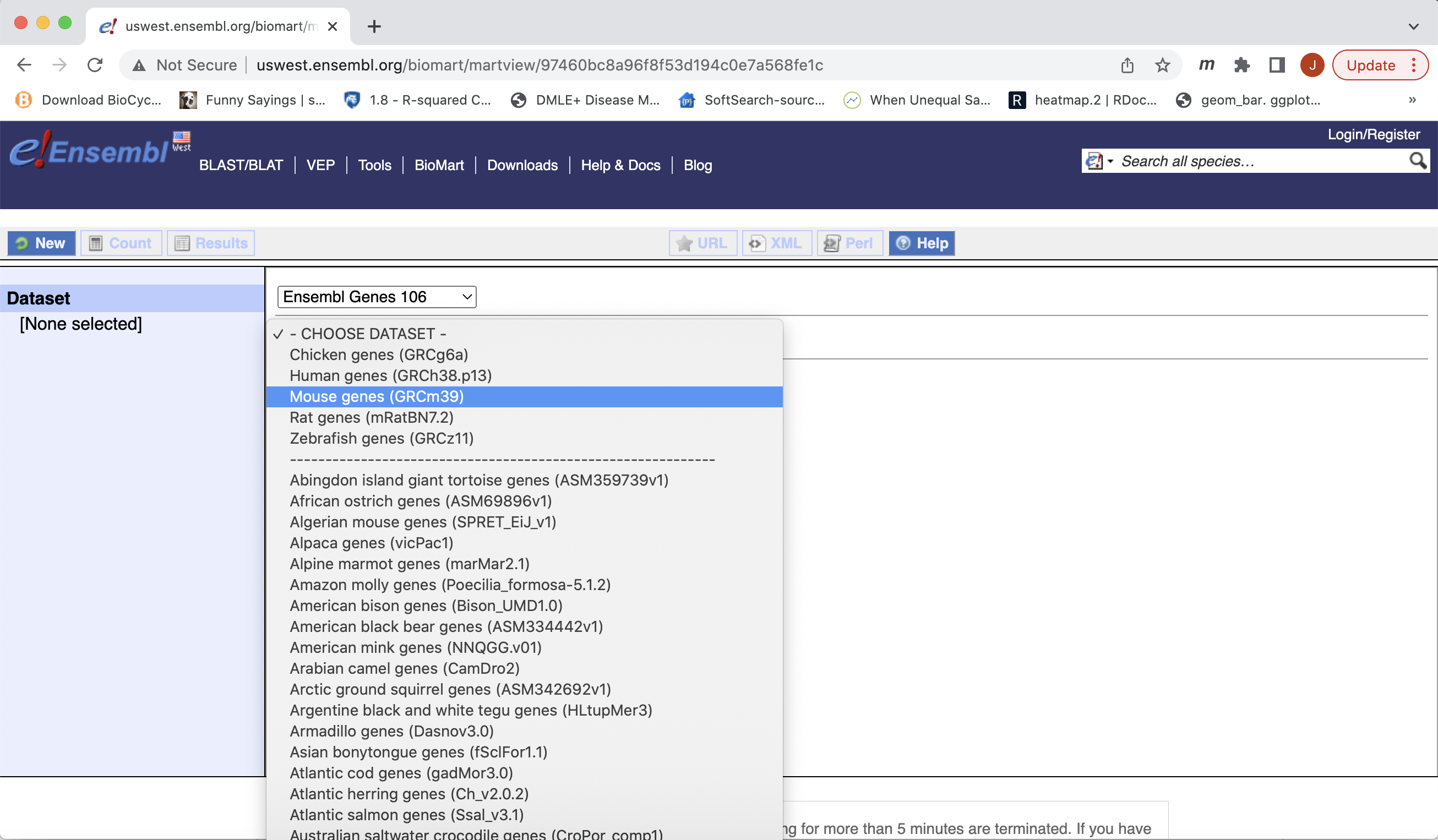
- Then the Organism, Here Mouse genes which is based on the GRCm39 genome.
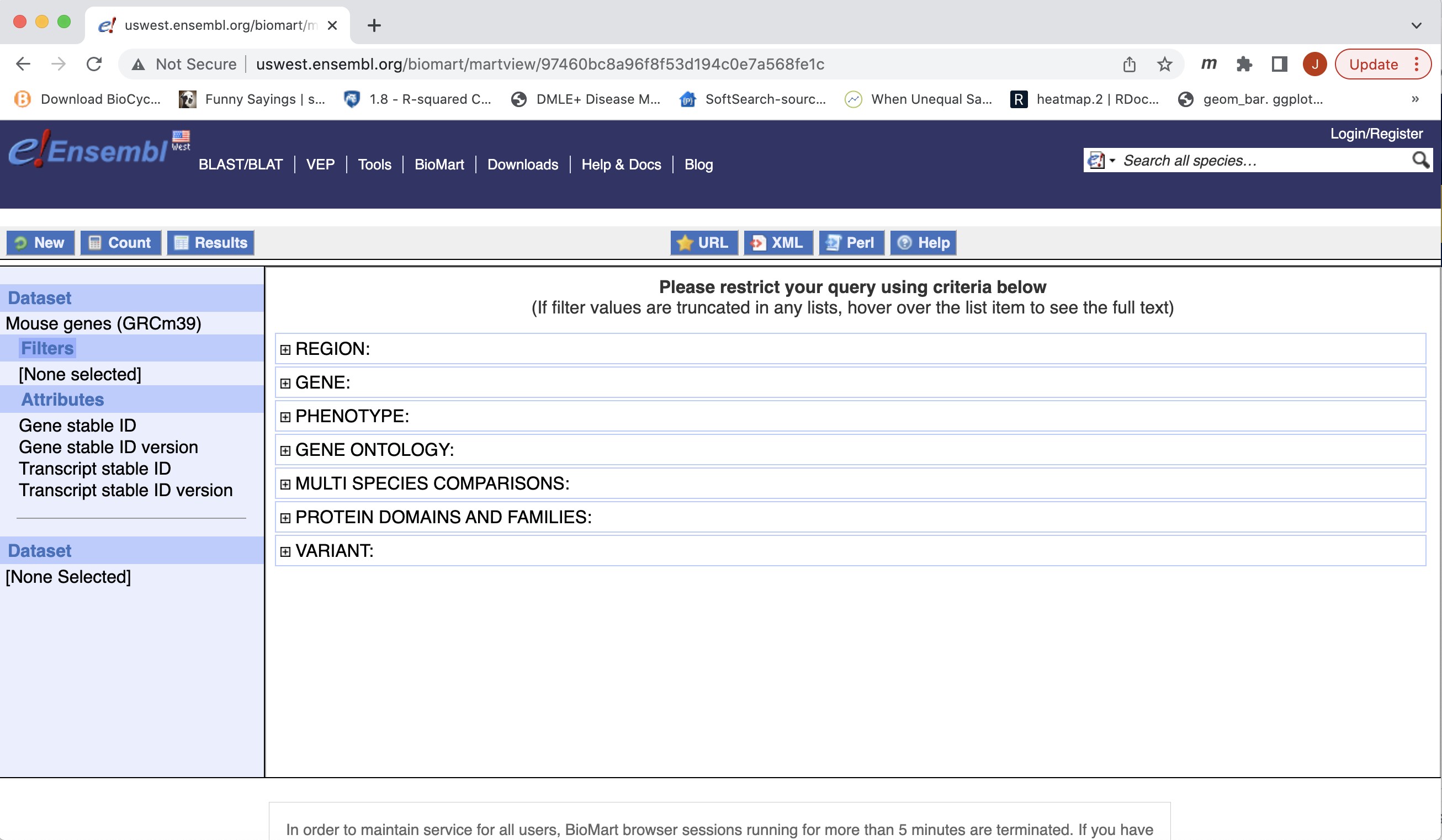
- You can choose to filter to only a subset of genes. Or a chromosome, or regions. We won’t filter here. BY default, all genes in the genome are selected.
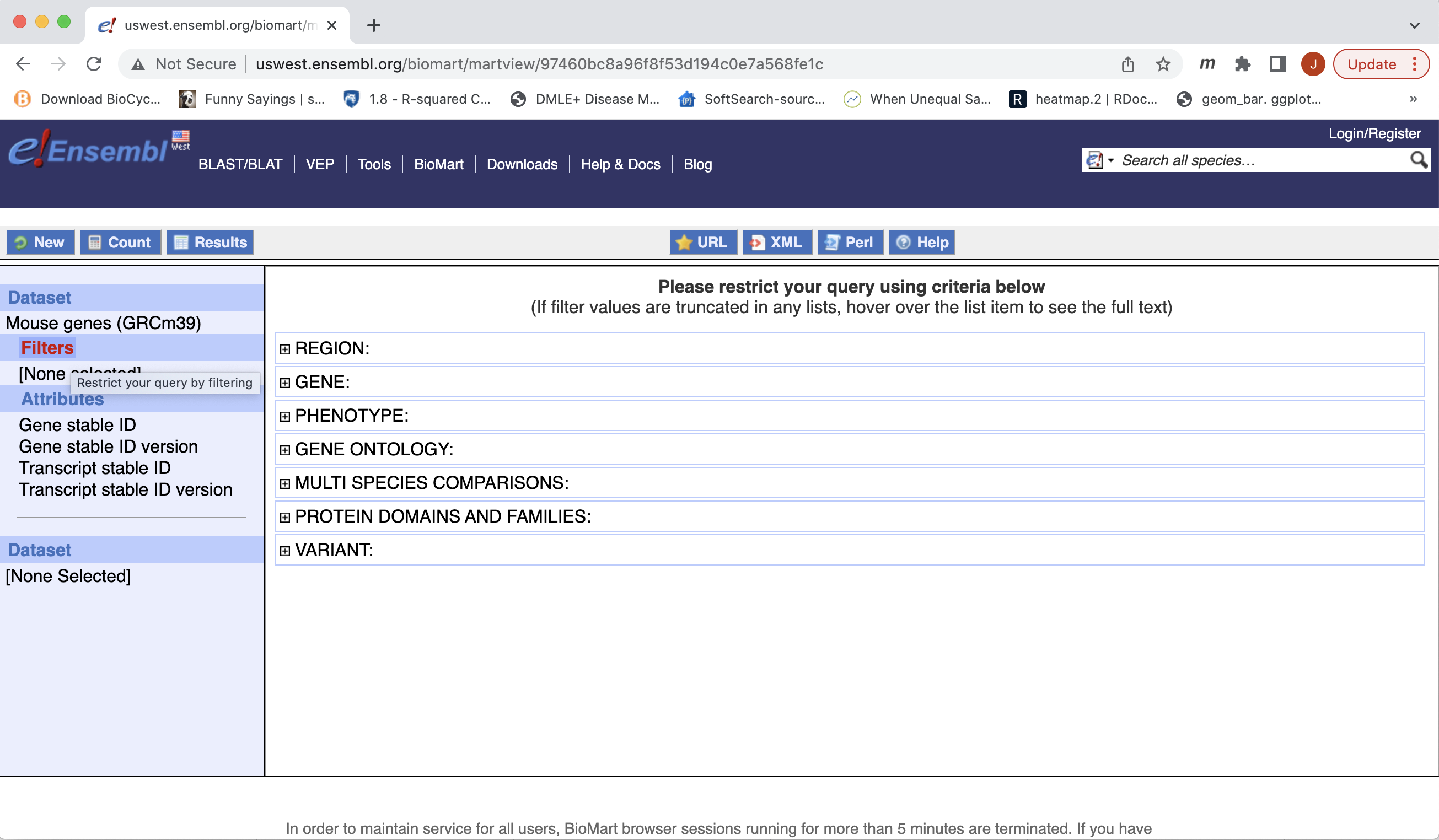
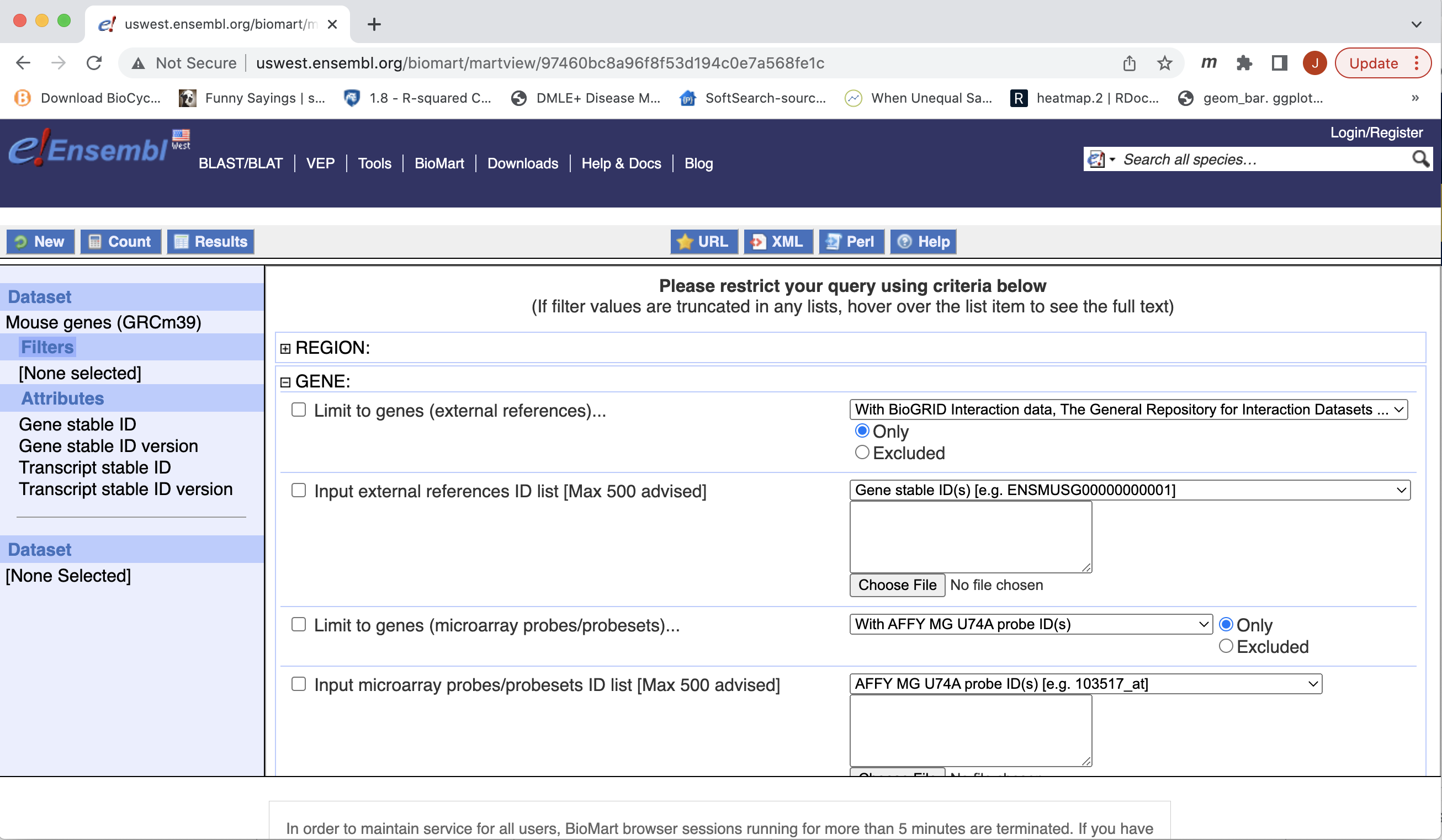
- Next select the attributes you want in the table.
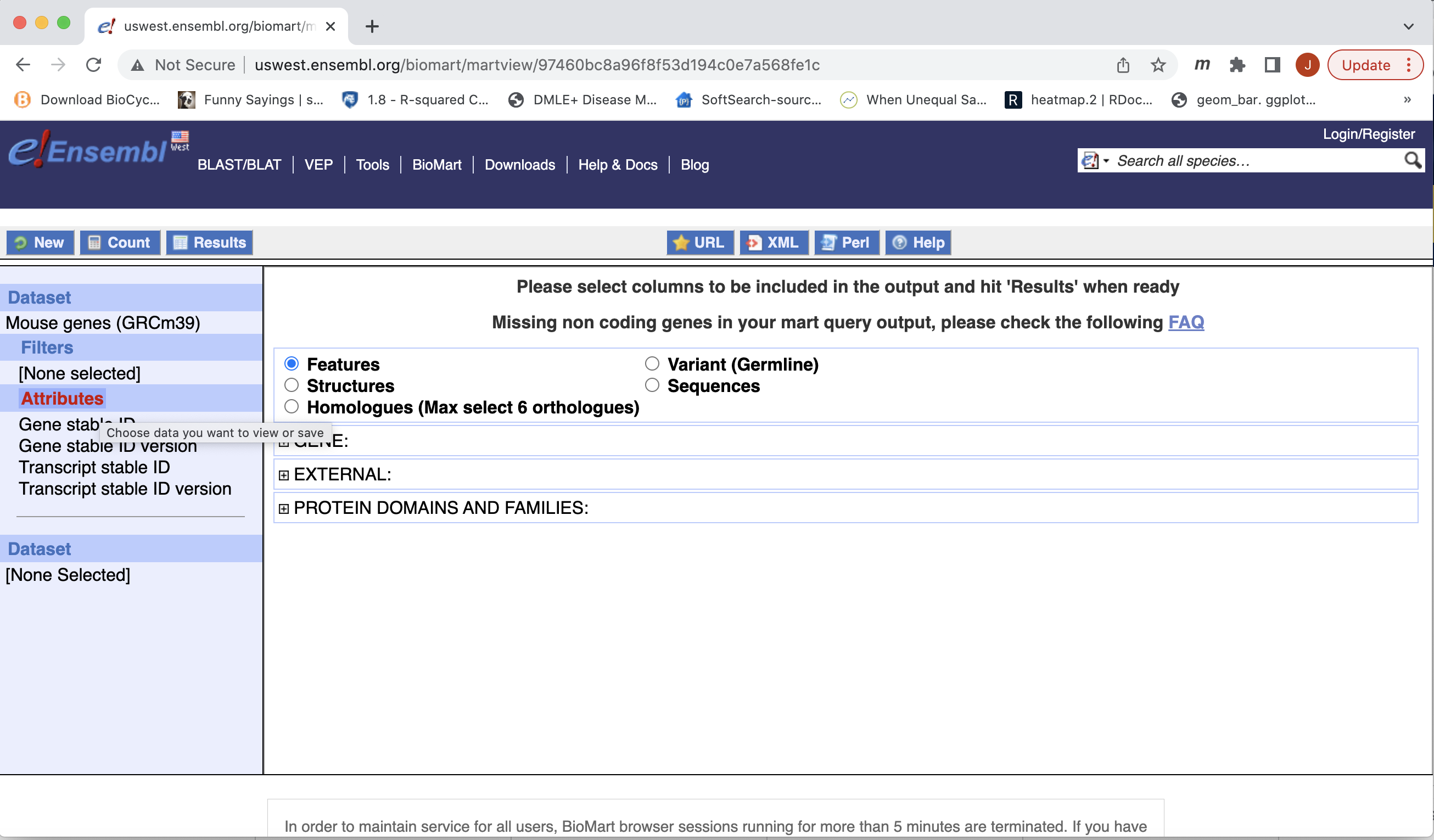
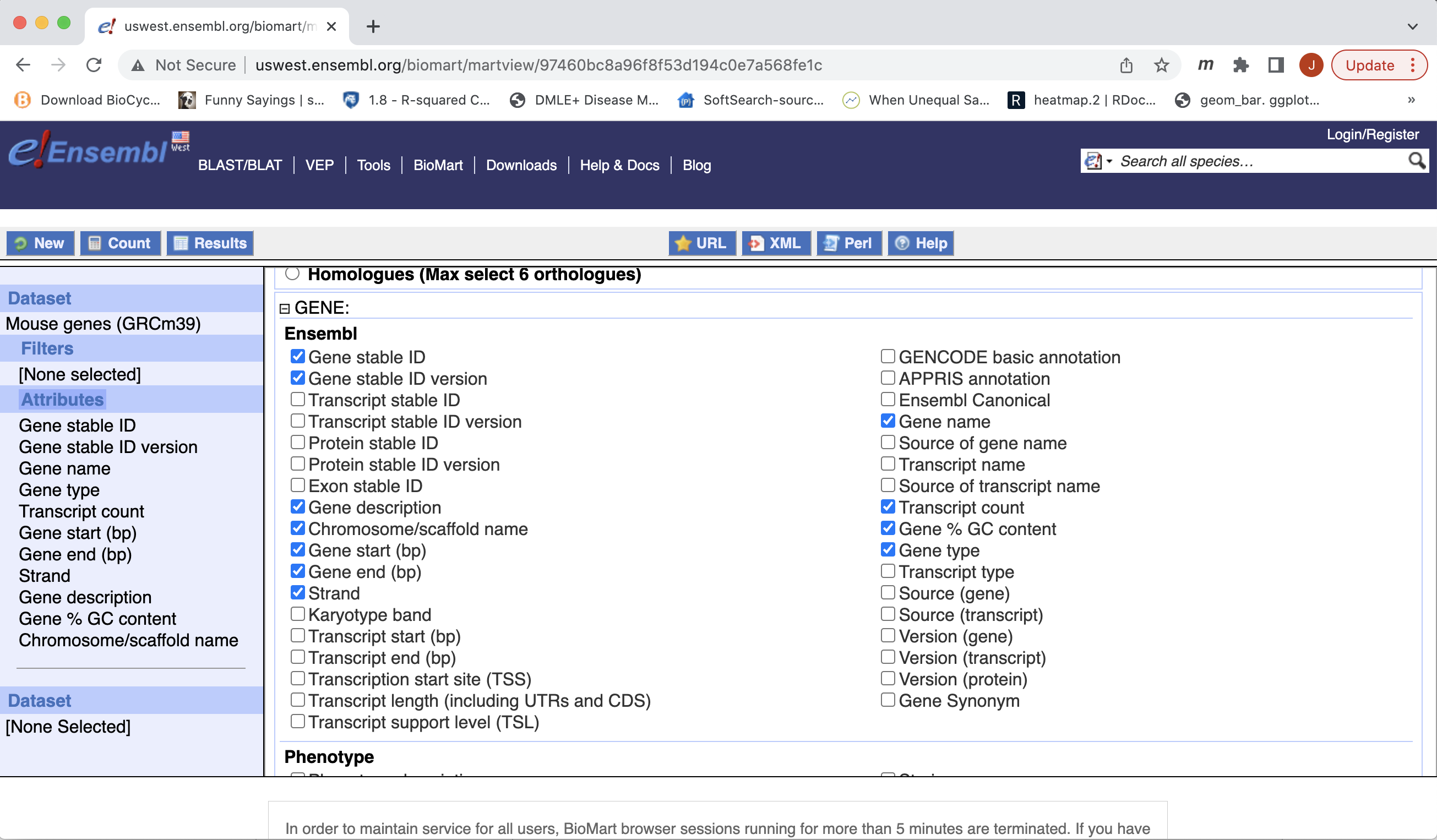
- Expand the ‘GENE’ tab, and select the attributes you want to retrieve. HERE recreate the list you see on the left side.
- Click “Results” (Top left -ish).
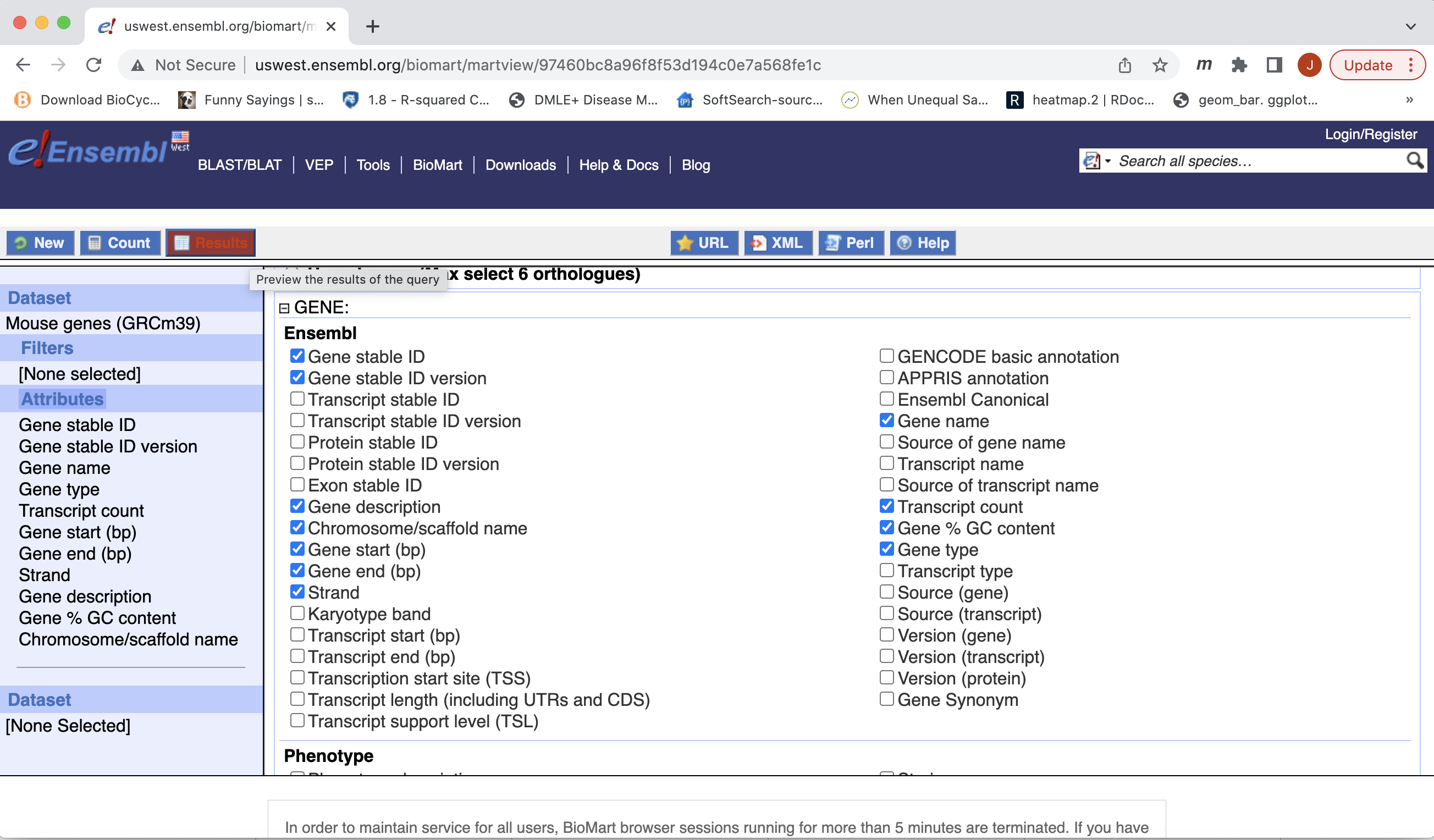
- Select “GO”, to download a tab-separated value (tsv) file.
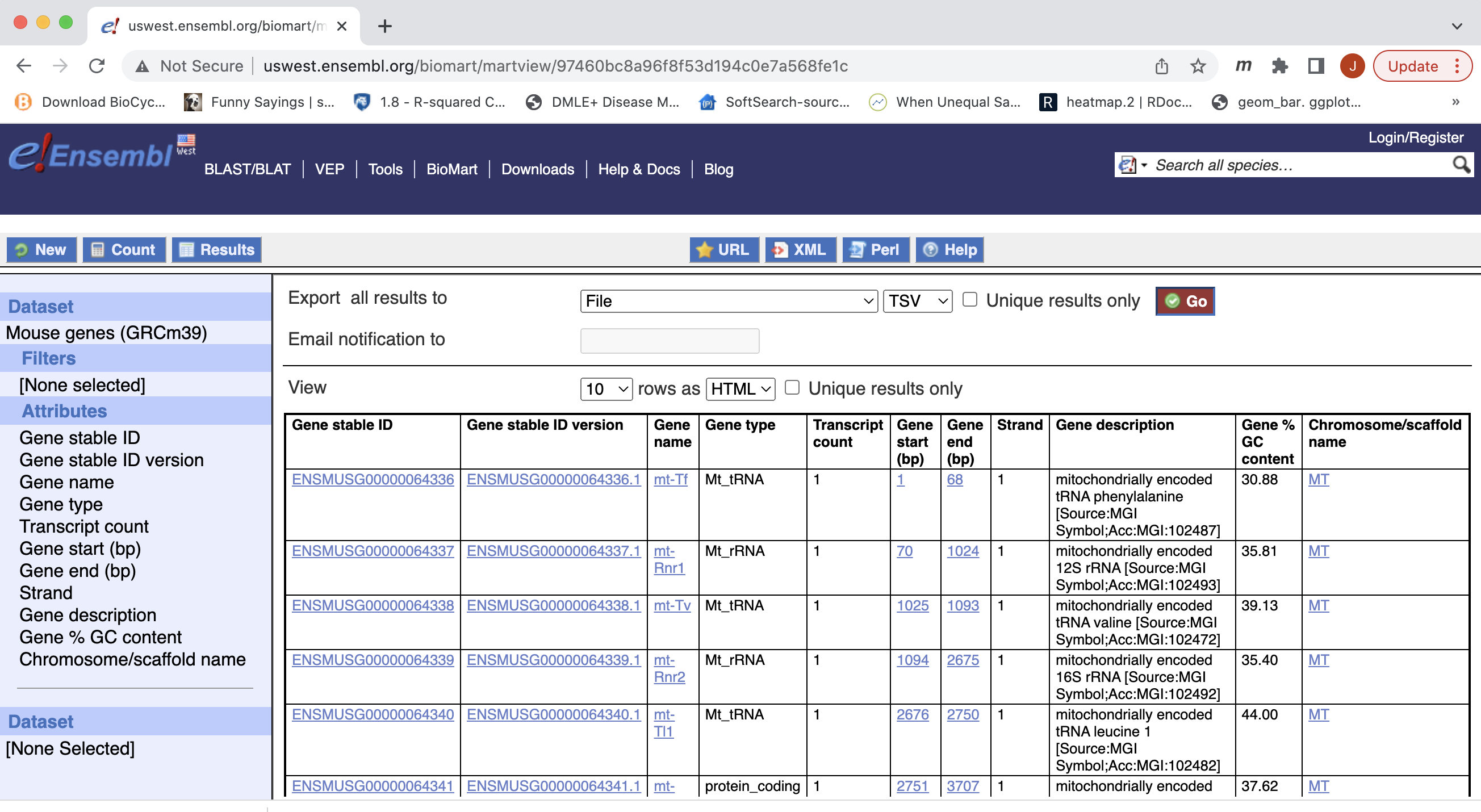
- The file will save as “mart_export.txt”, put the file into our working directory, rename to “ensembl_mm_106.tsv” and open the file in Excel to view the annotation.
