Some Graphical Summaries
Load our libraries
# Set up global options for nice reports and keeping figures:
knitr::opts_chunk$set(fig.width=14, fig.height=8, fig.align="center",
warning=FALSE, message=FALSE)
Lets start by loading libraries
library(phyloseq)
library(phangorn)
library(ggplot2)
nice_colors = c("#999999", "#E69F00", "#56B4E9","#e98756","#c08160","#5800e6", "#CDDC49", "#C475D3",
"#E94B30", "#233F57", "#FEE659", "#A1CFDD", "#F4755E", "#D6F6F7","#EB6D58", "#6898BF")
Load prior results
load(file=file.path("rdata_objects", "transformed_objects.RData"))
Plotting a Phylogenetic Tree
ps.class = tax_glom(ps.1, "Phylum")
plot_tree(ps.class, label.tips="Phylum", color="Group", size='abundance') + scale_color_manual(values=nice_colors[-1]) +
ggtitle("Phylum Level, phylogeny") + facet_wrap(.~Temp, ncol=1)
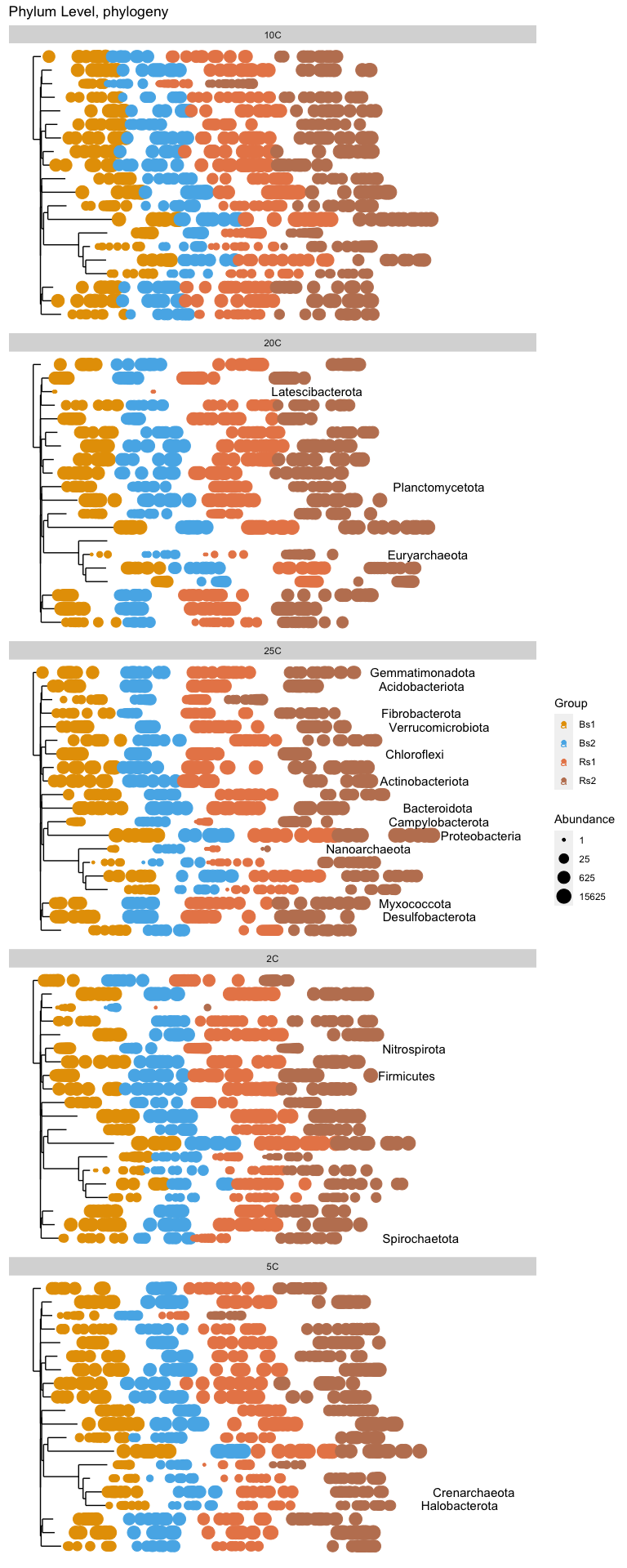
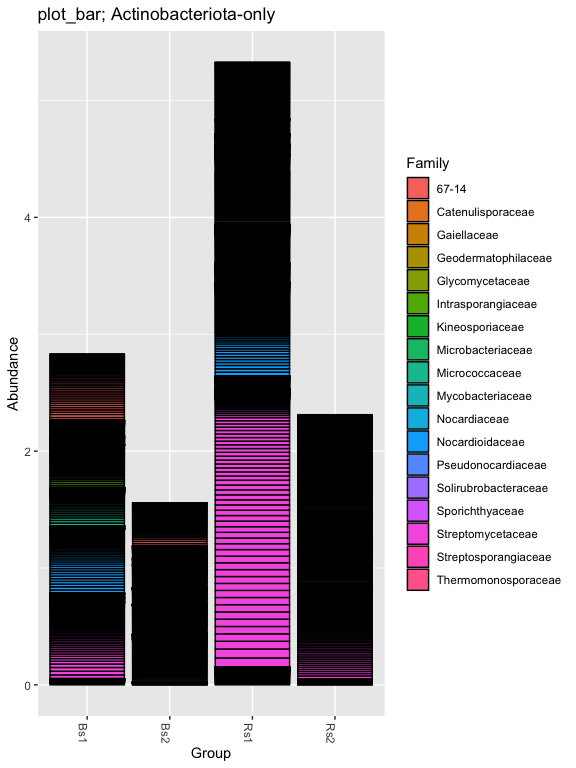
# Subset dataset by phylum
ps.1ra_acidob = subset_taxa(ps.1ra, Phylum=="Actinobacteriota")
plot_bar(ps.1ra_acidob, "Group", "Abundance", "Family", title= "plot_bar; Actinobacteriota-only")
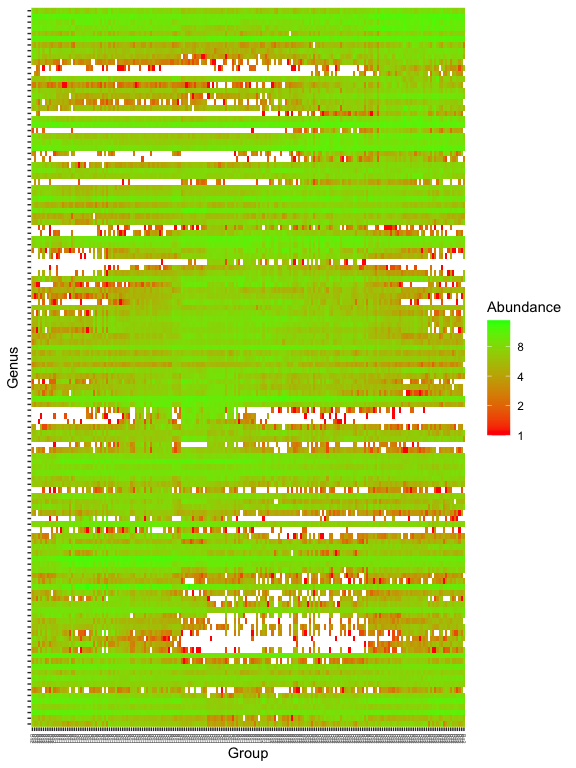
prop = transform_sample_counts(ps.1, function(x) x / sum(x) )
keepTaxa <- ((apply(otu_table(prop) >= 0.005,2,sum,na.rm=TRUE) > 2) | (apply(otu_table(prop) >= 0.05, 2, sum,na.rm=TRUE) > 0))
table(keepTaxa)
## keepTaxa
## FALSE TRUE
## 45 126
ps.1logRLE_trim <- prune_taxa(keepTaxa,ps.1logRLE)
plot_heatmap(ps.1logRLE_trim, "PCoA", distance="bray", sample.label="Group", taxa.label="Genus", low="red", high="green", na.value="white")
Cleanup
Get next Rmd
download.file("https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2021-May-Microbial-Community-Analysis/master/data_analysis/mca_part6.Rmd", "mca_part6.Rmd")
Record session information
sessionInfo()
## R version 4.0.3 (2020-10-10)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.3.3 phangorn_2.7.0 ape_5.5 phyloseq_1.34.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.6 lattice_0.20-44 prettyunits_1.1.1
## [4] Biostrings_2.58.0 assertthat_0.2.1 digest_0.6.27
## [7] foreach_1.5.1 utf8_1.2.1 R6_2.5.0
## [10] plyr_1.8.6 stats4_4.0.3 evaluate_0.14
## [13] highr_0.9 pillar_1.6.1 zlibbioc_1.36.0
## [16] rlang_0.4.11 progress_1.2.2 data.table_1.14.0
## [19] vegan_2.5-7 jquerylib_0.1.4 S4Vectors_0.28.1
## [22] Matrix_1.3-3 rmarkdown_2.8 labeling_0.4.2
## [25] splines_4.0.3 stringr_1.4.0 igraph_1.2.6
## [28] munsell_0.5.0 compiler_4.0.3 xfun_0.23
## [31] pkgconfig_2.0.3 BiocGenerics_0.36.1 multtest_2.46.0
## [34] mgcv_1.8-35 htmltools_0.5.1.1 biomformat_1.18.0
## [37] tidyselect_1.1.1 tibble_3.1.2 quadprog_1.5-8
## [40] IRanges_2.24.1 codetools_0.2-18 permute_0.9-5
## [43] fansi_0.4.2 withr_2.4.2 crayon_1.4.1
## [46] dplyr_1.0.6 MASS_7.3-54 rhdf5filters_1.2.1
## [49] grid_4.0.3 nlme_3.1-152 jsonlite_1.7.2
## [52] gtable_0.3.0 lifecycle_1.0.0 DBI_1.1.1
## [55] magrittr_2.0.1 scales_1.1.1 stringi_1.6.2
## [58] farver_2.1.0 XVector_0.30.0 reshape2_1.4.4
## [61] bslib_0.2.5.1 ellipsis_0.3.2 vctrs_0.3.8
## [64] generics_0.1.0 fastmatch_1.1-0 Rhdf5lib_1.12.1
## [67] iterators_1.0.13 tools_4.0.3 ade4_1.7-16
## [70] Biobase_2.50.0 glue_1.4.2 purrr_0.3.4
## [73] hms_1.1.0 parallel_4.0.3 survival_3.2-11
## [76] yaml_2.2.1 colorspace_2.0-1 rhdf5_2.34.0
## [79] cluster_2.1.2 knitr_1.33 sass_0.4.0
