Login to tadpole and navigate to your directory on the share space.
cd /share/workshop/adv_scrna/$USER
srun -t 1-00:00:00 -c 4 -n 1 --mem 16000 --partition production --account adv_scrna_workshop --reservation adv_scrna_workshop --pty /bin/bash
Immune profiling V(D)J T Cell and B Cell analysis with 10X
Human samples and mouse strains of types C57BL/6 and BALB/c have been tested within the 10X genomics system. If you use another mouse strain or different organism, then you need to create your own primers and reference sequence.
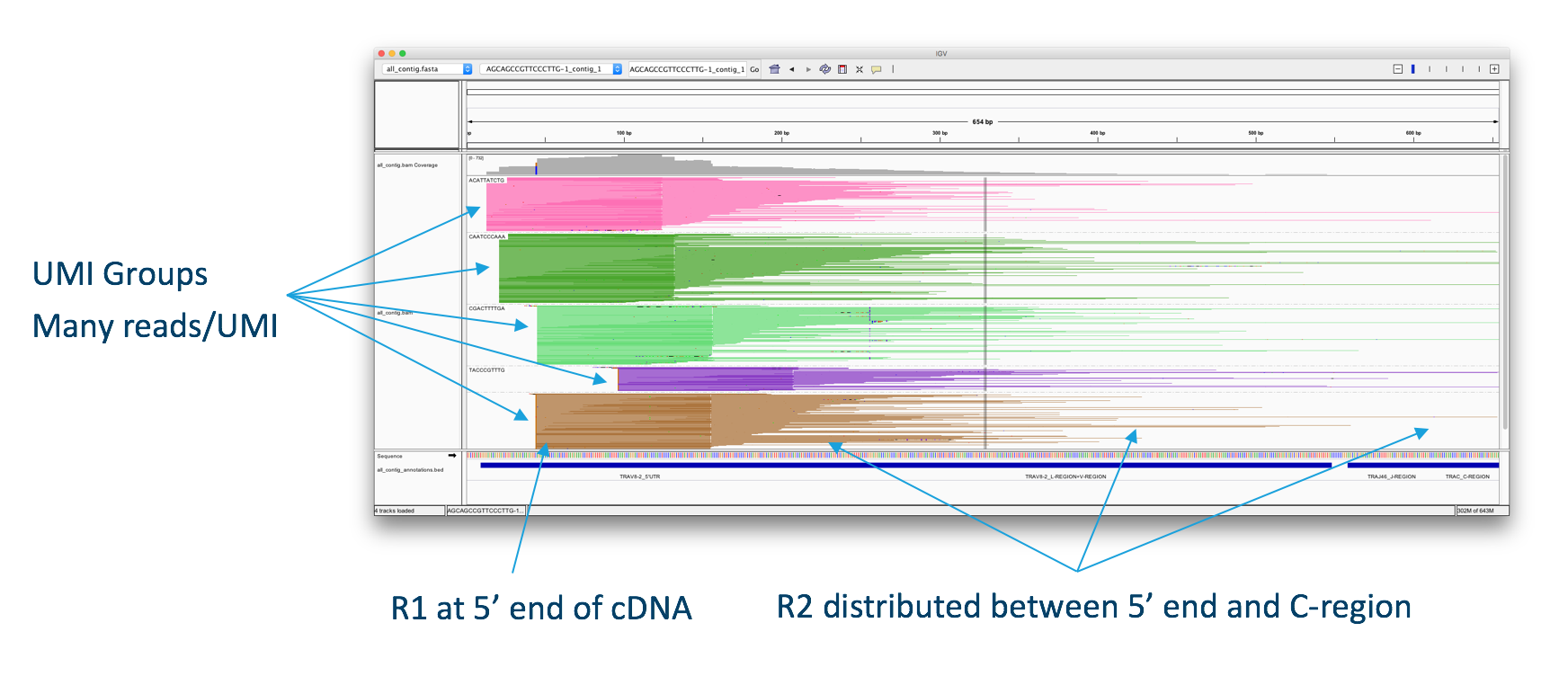
The V(D)J Algorithm
First V(D)J read-pairs aligned to an assembled contig, illustrating the structure of the read data. One to many UMIs are captured for each V(D)J chain. A round of enrichment PCR targeting the 5′ end to the C-region, followed by enzymatic fragmentation results in a pool of molecules originating from the same transcript. The molecules carry the same 10x barcode and UMI sequences, but with different insert lengths, resulting in different R2 start points. The diversity of R2 start points gives complete coverage of the targeted portion of each transcript, which is typically ~650bp.
Recommended sequencing depth is ~5,000 reads per cell.
Test dataset
The dataset we’ll be using is from the 10X website. Specifically we’ll be using the PBMCs from BALB/c mice dataset.
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing
mkdir cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/cellranger_vdj
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/cellranger_vdj
ln -s /share/biocore/workshops/2020_scRNAseq/VDJ/VDJ_output 00-RawData
Lets buld a reference for Mouse based on Ensembl release-100 VDJ entries
First lets setup a References folder for our experiment.
mkdir -p /share/workshop/adv_scrna/$USER/scrnaseq_processing/reference
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/reference
We should already have the needed genome and gtf file for Mouse Ensembl version 100
10X Genomics - cellranger vdj
Description of cellranger vdj can be found here
Building indexes for cellranger vdj (takes a long time)
10X Genomics provides pre-built references for human and mouse vdj regions to use with Cell Ranger. Researchers can make custom references for additional species or add custom vdj sequences of interest to the reference. The following tutorial outlines the steps to build a custom vdj reference using the cellranger mkvdjref pipeline and an ensembl genome.
You should already have the Ensembl 100 Mouse genome and GTF file in the reference folder, however if you don’t, you can copy then from here.
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/reference
ln -s /share/biocore/workshops/2020_scRNAseq/Reference/Mus_musculus.GRCm38.dna.primary_assembly.fa .
ln -s /share/biocore/workshops/2020_scRNAseq/Reference/Mus_musculus.GRCm38.100.gtf .
Running cellranger mkvdjref
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/reference
module load cellranger/3.1.0
cellranger mkvdjref \
--genome=GRCm38.cellranger_vdj \
--fasta=Mus_musculus.GRCm38.dna.primary_assembly.fa \
--genes=Mus_musculus.GRCm38.100.filtered.gtf \
--ref-version=3.1.0
This assumes that the following biotypes are present in the gtf files
- TR_C_gene
- TR_D_gene
- TR_J_gene
- TR_V_gene
- IG_C_gene
- IG_D_gene
- IG_J_gene
- IG_V_gene
You can also generate vdj references for IMGT sequences. Additional instructions for building VDJ references can be found here
Running the V(D)J piepline
cd /share/workshop/adv_scrna/$USER/scrnaseq_processing/cellranger_vdj
cellranger vdj \
--id=vdj_v1_mm_balbc_pbmc_b_sm \
--fastqs=00-RawData \
--sample=vdj_v1_mm_balbc_pbmc_b_sm \
--reference=../reference/GRCm38.cellranger_vdj
The V(D)J pipeline outputs alot of files
the output contains
- web_summary.html - similar to gene expression
- metrics_summary.csv - similar to gene expression
- annotation CSV/JSONs - filtered_contig_annotations.csv, clonotypes.csv
- FASTQ/FASTAs - filtered_contig.fasta/filtered_contig.fastq
- barcoded BAMs - consensus alignment mapping files
- cell_barcodes.json - barcodes which are identified as targeted cells.
