Quick Introduction to the Workshop
The mission of the Bioinformatics Core facility is to facilitate outstanding omics-scale research through these activities:

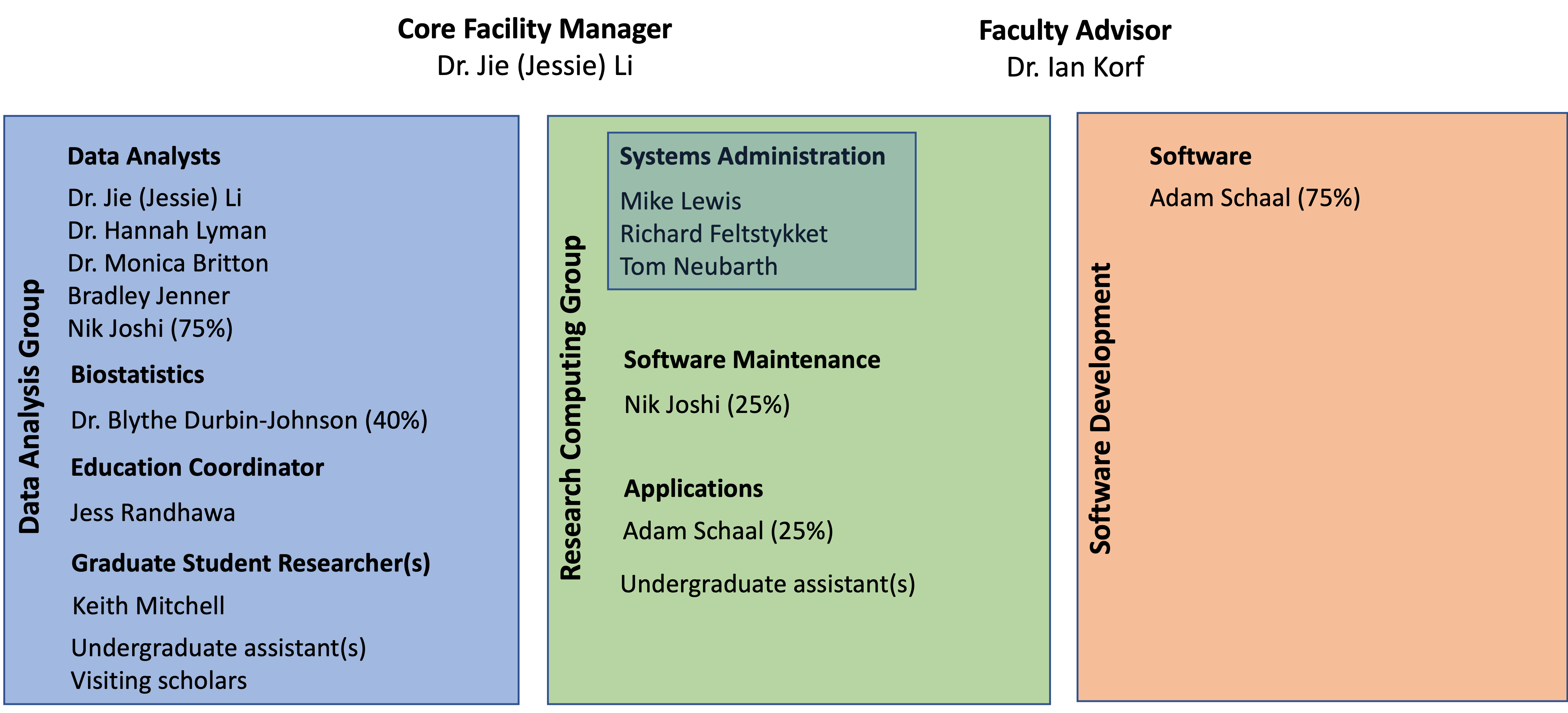
Bioinformatics Core Staff and Students
Our team offers custom bioinformatics services to academic and private organizations. We have a strong academic background with a focus on cutting edge, open source software. We replicate standard analysis pipelines (best practices) when appropriate, and/or develop novel applications and pipelines when needed, however we always emphasize biological interpretation of the data.
Collaboration with PacBio, Richard Kuo, and Conesa Lab
This workshop prominently features people from PacBio, the Roslin Institute, and Ana Conesa’s lab from the University of Florida. From PacBio Dr. Liz Tseng is a primary developer of much of (if not all of) PacBio’s Isoseq pipeline, Richard Kuo is a bioinformatician at the Roslin Institute and the creator and developer of the TAMA software package for transcriptome annotation of non-model organisms, and Dr. Ana Conesa and her lab are most notable for their software Blast2GO, maSigPro, Paintomics, NOISeq, SQANTI and tappAS, lab website (faculty page).


UCD Bioinformatics Core Contacts
-
Request for data analysis services, consultations.
-
Bioinformatics training course information
-
Computing Issues, include but not limited to user account questions, equipment failure/malfunction, software install, software failures (not related to use).
Structure of the Virtual Workshops
Given the current Covid19 pandemic, and stay in place orders around the world, we’ve decided to take our popular bioinformatic workshops virtual. Our intent is to offer as close to an experience we can to our in-person workshops. We will hold the same goals and strive for a similar lecture/hand-on ratio. We will be using multiple technologies in order to help facilitate a maximum amount of interaction.
Zoom
Course lecture, discussions, and one-on-one help/troubleshooting will be conducted using a zoom meeting.
- Each participant will be provided a personal break out room that will allow one-on-one troubleshooting by our staff, screen sharing and even remote control.
- The Chat features of Zoom will not be our primary mode of text communication as its history is not reliable.
- We will stick to a schedule to allow people to plan their day and know when they need to be back at their computers for lecture.
- Video recordings of lectures will be made available to participants.
Because video is involved, we ask everyone to be respectful and we reserve the right to remove someone if they are being disrespectful or disruptive.
Learn more about how we use Zoom in our workshops.
Slack
Text based communication will be conducted via a Slack channel. Staff will be monitoring the Slack channel to answer questions (and schedule a Zoom break out room if needed). If you know the answer to someone else’s question, feel free to answer it.
Learn more about how we use Slack in our workshops.
Patience
We ask for a little more patience as this process is still new to us, but we are committed to providing you the best educational experience we can (under the current conditions).
Workshop Goals
- End to End understanding of the basics of Iso-Seq experiments
- Lectures/Discussions
- Technology
- Experimental design
- Cost estimation
- Workflow
- Data reduction
- Data analysis
-
To work through a complete experiment, starting from raw data to completion, including making a few figures.
- Goal is 30-40% lecture/discussion 60-70% hands-on
Workshop Materials
Workshop materials are all posted on github, and publicly available
http://bioinformatics.ucdavis.edu/training
-
Workshop registration site:
-
Public Github repository listing:
-
This Isoseq workshop
https://ucdavis-bioinformatics-training.github.io/2020-september-isoseq/
Computing Cluster
A portion of this course will be conducted on our servers and compute cluster (tadpole.genomecenter.ucdavis.edu).
Everyone should get an account.
https://computing.genomecenter.ucdavis.edu
Request an account -> sponsor is “Bioinformatics Core Workshop”
If you already have, or have had in the past, an account on our systems, then please tell us your username, or email, and we will add you to the workshop. Do not create a new account
Cluster usage will be under the slurm reservation ‘isoseq_workshop’
