Closing Thoughts
The forest through the trees.
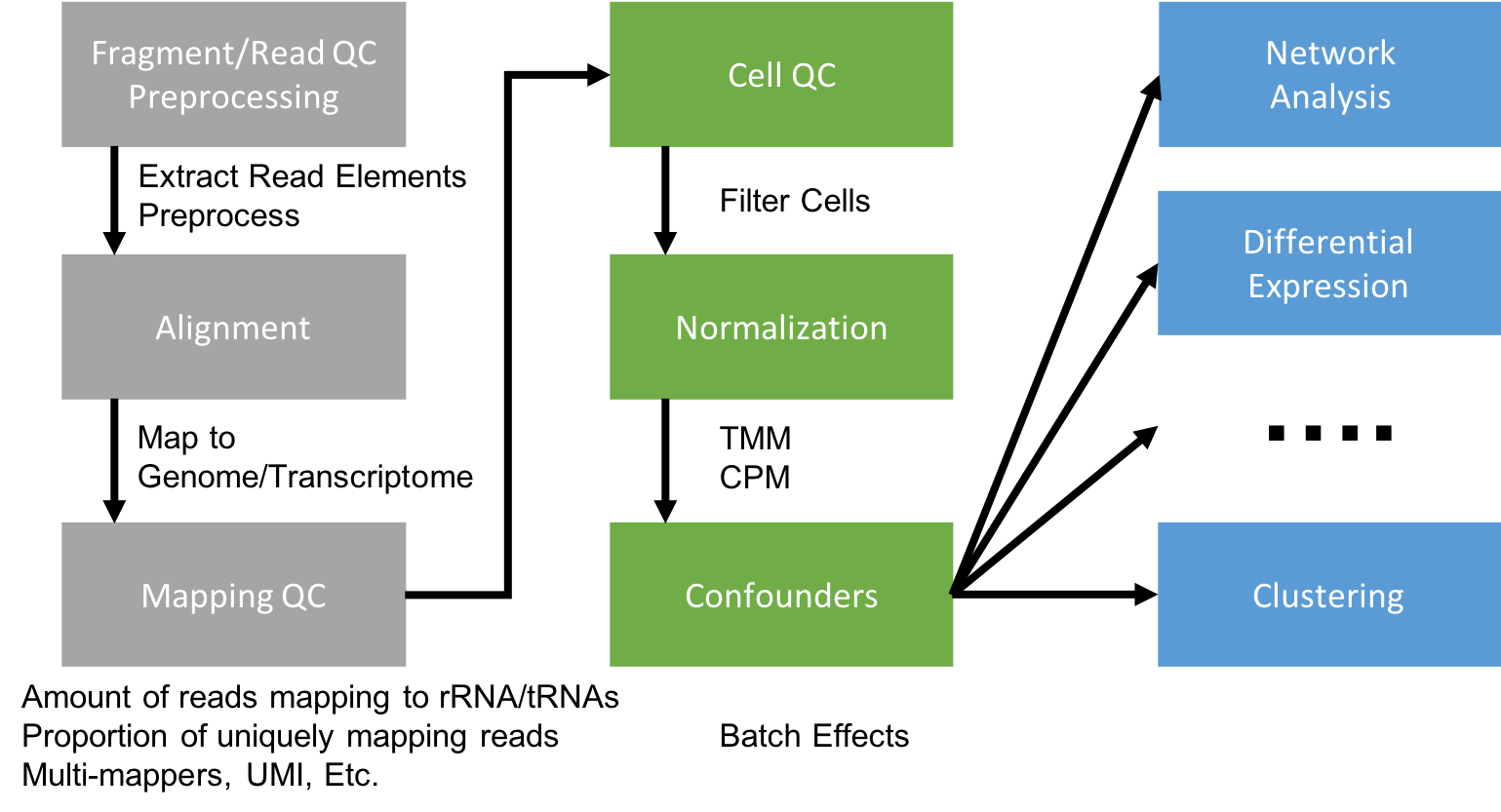
Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
- Be Consistent
Prerequisites for bioinformatics
- Access to a multi-core (24 cpu or greater), ‘high’ memory 64Gb or greater Linux server.
- Familiarity with the ’command line’ and at least one programming language.
- Basic knowledge of how to install software
- Basic knowledge of R (or equivalent) and statistical programming
- Basic knowledge of Statistics and model building
The bottom line
- Spend the time (and money) planning and producing good quality, accurate and sufficient data for your experiment.
- Get to know to your data, develop and test expectations
- Result, you’ll spend much less time (and less money) extracting biological significance and results during analysis.
Next Steps
My recommendation is to follow all of the instructions again, from the beginning on your own and send emails to
training.bioinformatics@ucdavis.edu
And we will be responsive to answering questions
Upcoming Workshops
Now in our 12th year of training researchers, the UC Davis Bioinformatics Core invites you to attend one or more of our 2019 workshops. The following workshops are suitable for beginners, and are also of interest to those with some data analysis experience.
For more information on these and other events, and to register, go to https://registration.genomecenter.ucdavis.edu/
December 16-19: Bioinformatics Prerequisites Workshop @ UC Davis
This workshop will cover the basic computational and statistical concepts needed before performing bioinformatics analysis.
- Access to a multi-core (24 cpu or greater), ‘high’ memory 64Gb or greater Linux server.
- Familiarity with the ’command line’ and constructing scripts/pipelines.
- Basic knowledge of how to install software
- Basic knowledge of R (or equivalent) and statistical programming
- Basic knowledge of Statistics and model building
The course will include experimental data organization, basic command line and high performance computing concepts, how to install software, use help, run applications, and the basics of building scripts and pipelines. Also covered will be basic R programming, working with data tables and generating figures, basic statistical concepts, and statistical model building. There are no prerequisites for this workshop other than an interest in bioinformatics! The workshop will be on the UC Davis campus, and will run from 9am to 4:30pm each day and include light breakfast, lunch, and snacks.
The registration fee for the entire workshop is $600 for UC and UC affiliates, $900 for other academic or non-profit participants and $1,200 for other participants. We accept credit cards, as well as UC Davis recharge accounts, for payment. Registration fees include light breakfast, lunch, and snacks, but do not include dinner, lodging or parking fees.
UNIX/COMMAND LINE CONCEPTS
- Introductory materials, Unix/bash command line basics.
- Environment variables, your bash profile
- Basic bash shell scripting.
- Introduction to cluster computing
- Basics of git
- Folder and experiment structure
- Installation and running of simple sequencing tools.
- Constructing and run a simple bioinformatics pipeline.
R/DATA PROGRAMMING CONCEPTS
- Introduction to the R programming language.
- R markdown and notebooks.
- Constructing metadata tables, data munging, excel to R
- Basics of the tidyverse, data frame manipulation (join/rename/pipes)
- Figures in R, ggplot2
- Statistical concepts and model building.
If you would like to take a look at the documentation from previous workshops, see https://bioinformatics.ucdavis.edu/training/documentation/.
Questions? Contact us at training.bioinformatics@ucdavis.edu Email List Note: If you received this email directly from training.bioinformatics@ucdavis.edu, you are currently subscribed to one of our announcements lists. To subscribe / unsubscribe from the Bioinformatics Core email lists, please follow the instructions at the bottom of http://bioinformatics.ucdavis.edu/contact-us/.
