What is Tidyverse?

The tidyverse packages are developed and maintained by RStudio Inc.
- Also make RStudio, Rstudio Server, Shiny, and a variety of other tools.
- Open source with commercial products available.

Many publications including:
- Wickham, Hadley. “Tidy data.” Journal of Statistical Software 59.10 (2014): 1-23.
- Wickham, Hadley. “An introduction to ggplot: an implementation of the grammar of graphics in R.” Statistics (2006).
- Wickham, Hadley. “ggplot2.” Wiley Interdisciplinary Reviews: Computational Statistics 3.2 (2011): 180-185.
- Wickham, Hadley. “Reshaping data with the reshape package.” Journal of statistical software 21.12 (2007): 1-20.
Why was tidyverse invented?
The tidyverse was invented in response to a sense of frustration with the difficulty of performing data analysis using various combinations of existing R packages, which may require managing inconsistent syntax and naming schemes. The tidyverse packages focus on making the initial steps of data analysis, particularly organizing and manipulating data, easier.
Learn more about the frustrations and philosophy behind the origin of the tidyverse.
The tidyverse packages are very popular, but not everyone agrees that tidyverse is a great idea. Some of these concerns have been outlined in the TidyverseSkeptic document, these include:
- Tidyverse makes learning harder, due to adding much complexity leading to cognitive overload
- tidyverse tends to be verbose, with some packages containing hundreds of functions
- Adoption of Tidyverse may give Rstudio too much control of the open source R project
- RStudio provides frequent training sessions and webinars, which may result in new users not being exposed to alternatives
- Widespread adoption of Tidyverse may limit adoption of technologically superior packages in the future
While users don’t have to choose between tidyverse and base R, many people have picked a camp. You can find out how and why people pick sides:
- https://www.r-bloggers.com/why-i-dont-use-the-tidyverse/
- https://github.com/matloff/TidyverseSkeptic
- https://www.r-bloggers.com/the-tidyverse-curse/
- http://varianceexplained.org/r/teach-tidyverse/
- https://ds4ps.org/2019/04/20/datatable-vs-dplyr.html
Why are we learning tidyverse?
Although there are many alternatives, we’ll be spending time on learning tidyverse packages in this workshop because data science and tidyverse are increasingly intertwined.
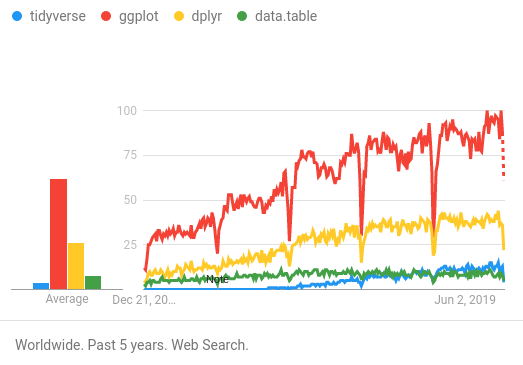
By familiarizing yourself with the tidyverse, you “future proof” yourself. If the next generation of R users are mostly tidyverse users, knowing tidyverse is a marketable skill, and performing analyses using tidyverse packages may improve the availability of code examples and help for the types of projects that interest you.
Getting started
In the R console run the following commands to ensure that you have packages installed:
if (!any(rownames(installed.packages()) == "knitr")){
install.packages("knitr")
}
if (!any(rownames(installed.packages()) == "tidyverse")){
install.packages("tidyverse")
}
library(knitr)
library(tidyverse)
Download the template Markdown workshop document and open it
In the R console run the following command:
download.file("https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2020-Bioinformatics_Prerequisites_Workshop/master/Intro_to_R/Intro2R/Intro_to_tidyverse_and_ggplot2.Rmd", "Intro_to_tidyverse_and_ggplot2.Rmd")
